Section summary #
Here I attempted to prepare 1 replicate of T7Init Mix 1 with different enzymatic
treatments. Fred pointed out that I should use chloroquine gels to confirm
topological manipulations where successful. Repeated chloroquine gels showed
Topoisomerase treatments to be effective at relaxing the plasmid mix but
Gyrase left something to be desired compared to Stoltz et al. Since
Gyrase was causing headaches I decided to drop it from this first go around
of sequencing.
During my first attempt with these samples bisulfite treatment and PCR amplification succeeded but I lost basically all of my samples at the agarose gel extraction step. I re-did the PCR reaction with my remaining bisulfite converted samples but this time the PCR failed for unknown reasons.
Ultimately this replicate yielded no usable samples.
Substrate preparations #
3/27/28 #
Four samples were prepared from T7InitMix-0 which contains approximately
10 ng / ul / construct. Volumes of each construct added were calculated
based on appearance of 1 ul of construct on agarose gel compared to pFC53 and
pFC9 (see experiment details here).
The four samples and described in the section below.
1: BamHI linearized #
| Reagent | Volume (ul) |
|---|---|
| npH20 | 80 |
| T7InitMix-1 | 8 |
| NEB BamHI-HF | 2 |
| NEB rCutSmart 10X Buffer | 10 |
Incubated 37C 1 hour.
2: Hyper-supercoiled (Gyrase treated) #
| Reagent | Volume (ul) |
|---|---|
| npH20 | 38 |
| T7InitMix-1 | 8 |
| Topogen 5x Gyrase Assay Buffer | 12 |
| TopoGen Gyrase Enzyme | 2 |
Incubated 37C 1 hour.
3: Relaxed (Topoisomerase I treated) #
| Reagent | Volume (ul) |
|---|---|
| npH20 | 80 |
| T7InitMix-1 | 8 |
| NEB 10x rCutSmart Buffer | 10 |
| NEB TopoisomeraseI | 2 |
Incubated 37C 20 mins.
After completing incubations all samples were phenol/chloroform extracted using homemade phase lock columns and then EtOH precipitated and re-suspended in 20 ul 10mM Tris-HCl. Samples were then stored at -20C.
Reagent details #
Reagent cat and lot numbers for the reactions described above are listed in the table below.
Nanodrop results #
After completing all treatments and precipitations nanodroped samples to confirm still have DNA.
| Sample | ng/ul | 260/280 | 260/230 |
|---|---|---|---|
| 0. Supercoiled | 267.5 | 1.8 | 2.035 |
| 1. Linearized | 357.2 | 1.8 | 2.06 |
| 2. Hyper-supercoiled | 290.5 | 1.732 | 1.772 |
| 3. Relaxed | 268.3 | 1.745 | 2.067 |
IVT with T7 and bisulfite conversion #
3/28/22 #
Prior to IVT all samples were spiked with 40 ng of pFC9 as a positive control. Samples were then treated according to IVT + Bisulfite treatment protocol.
Sample 0 was treated according to “IVT followed by bisulfite conversion for SMRF-seq samples requiring untranscribed controls” while others were treated according to “IVT followed by bisulfite conversion for SMRF-seq samples not requiring untranscribed controls”.
Sample table #
Following samples described in the table below were produced. Included along with nanodrop results after completing bisulfite conversion.
Also available in this document.
Sample table and nanodrop results
| Sample Series | Sample Number | Formal name | Transcribed | TopoisomeraseI | Gyrase | BamHI-HF | RnaseA | RnaseH | Deprotienzied | ng/ul | 260/280 | 260/230 | Name |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHS0 | 0 | EHS0-R0 | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | 3.6 | 1.16 | 0.457 | T7 SC Txn - |
| EHS0 | 1 | EHS0-R1 | TRUE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | 39 | 2.019 | 2.202 | T7 SC Txn + |
| EHS0 | 2 | EHS0-R2 | TRUE | FALSE | TRUE | FALSE | FALSE | FALSE | FALSE | 27.8 | 2.17 | 1.057 | T7 Gyr. Txn+ 2 |
| EHS0 | 3 | EHS0-R3 | TRUE | FALSE | TRUE | FALSE | FALSE | FALSE | FALSE | 15.6 | 3.395 | 3.756 | T7 Gyr. Txn+ 1 |
| EHS0 | 4 | EHS0-R4 | TRUE | TRUE | FALSE | FALSE | FALSE | FALSE | FALSE | 29.2 | 2.214 | 2.133 | T7 Topo Txn+ 1 |
| EHS0 | 5 | EHS0-R5 | TRUE | TRUE | FALSE | FALSE | FALSE | FALSE | FALSE | 29.9 | 2.62 | 2.006 | T7 Topo Txn+ 2 |
| EHS0 | 6 | EHS0-R6 | TRUE | FALSE | FALSE | TRUE | FALSE | FALSE | FALSE | 49.3 | 2.117 | 2.372 | T7 Lin. Txn+ 1 |
| EHS0 | 7 | EHS0-R7 | TRUE | FALSE | FALSE | TRUE | FALSE | FALSE | FALSE | 48.7 | 2.196 | 1.446 | T7 Lin. Txn+ 2 |
| EHS0 | 8 | EHS0-R8 | TRUE | FALSE | FALSE | FALSE | TRUE | FALSE | TRUE | 5.4 | 1.387 | 0.453 | T7 DP SC Txn+ |
| EHS0 | 9 | EHS0-R9 | TRUE | FALSE | TRUE | FALSE | TRUE | FALSE | TRUE | 1.3 | 0.713 | 0.138 | T7 DP Gyr. Txn+ 1 |
| EHS0 | 10 | EHS0-R10 | TRUE | FALSE | TRUE | FALSE | TRUE | FALSE | TRUE | 9.4 | 3.22 | 1.467 | T7 DP Gyr. Txn+ 2 |
| EHS1 | 11 | EHS1-R11 | TRUE | TRUE | FALSE | FALSE | TRUE | FALSE | TRUE | -0.5 | -0.486 | 0.988 | T7 DP Topo. Txn+ 1 |
| EHS2 | 12 | EHS2-R12 | TRUE | TRUE | FALSE | FALSE | TRUE | FALSE | TRUE | -0.2 | -0.275 | -0.032 | T7 DP Topo. Txn+ 2 |
| EHS3 | 13 | EHS3-R13 | TRUE | FALSE | FALSE | TRUE | TRUE | FALSE | TRUE | 0.6 | 0.543 | 0.084 | T7 DP Lin. Txn+ 1 |
| EHS4 | 14 | EHS4-R14 | TRUE | FALSE | FALSE | TRUE | TRUE | FALSE | TRUE | 2.6 | 1.336 | 0.344 | T7 DP Lin. Txn+ 2 |
| EHS5 | 15 | EHS5-R15 | TRUE | FALSE | FALSE | FALSE | TRUE | TRUE | TRUE | -4.6 | -0.482 | -0.124 | T7 DP SC RH1 Txn+ |
While sample values were quite low (even for bisulfite) I think it is still worth moving forward with the PCR reactions to really see if anything is in those tubes. What was surprising was that proteinized samples tended to have much higher specs. This may have been due to the lack of RnaseA digestion with these samples though.
Reagent details #
Transcription and Bisulfite conversion reagents
| Reagent | Cat Number | Lot Number | Notes |
|---|---|---|---|
| Zymogen lightning conversion reagent | D5030-1 | 211521 | Used for treating protienized transcribed samples and supercoiled un-transcribed control |
| Zymogen lightning conversion reagent | D5030-1 | 211521 | Used for treating de-protienized samples |
| Zymogen EZ DNA Methylation lightning kit | 11-338 | ZRC202947 | |
| NEB RnaseH buffer 10x | B0297S | 10085445 | |
| NEB RNA Pol Reaction Buffer 10x | B9012S | 10073286 | |
| Thermo Scientific RNA grade glycogen | R0551 | 1191453 | |
| NEB rNTP Mix | N0466S | 10085868 |
3/29/22 #
PCR of bisulfite converted samples #
| Formal name | Name | Transcribed | TopoisomeraseI | Gyrase | BamHI-HF | RnaseA | RnaseH | Deprotienzied | ng/ul | 260/280 | 260/230 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EHS0-R0 | T7 SC Txn - | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | 3.6 | 1.16 | 0.457 |
| EHS0-R1 | T7 SC Txn + | TRUE | FALSE | FALSE | FALSE | FALSE | FALSE | FALSE | 39 | 2.019 | 2.202 |
| EHS0-R6 | T7 Lin. Txn+ 1 | TRUE | FALSE | FALSE | TRUE | FALSE | FALSE | FALSE | 49.3 | 2.117 | 2.372 |
| EHS0-R7 | T7 Lin. Txn+ 2 | TRUE | FALSE | FALSE | TRUE | FALSE | FALSE | FALSE | 48.7 | 2.196 | 1.446 |
| EHS0-R8 | T7 DP SC Txn+ | TRUE | FALSE | FALSE | FALSE | TRUE | FALSE | TRUE | 5.4 | 1.387 | 0.453 |
| EHS3-R13 | T7 DP Lin. Txn+ 1 | TRUE | FALSE | FALSE | TRUE | TRUE | FALSE | TRUE | 0.6 | 0.543 | 0.084 |
| EHS4-R14 | T7 DP Lin. Txn+ 2 | TRUE | FALSE | FALSE | TRUE | TRUE | FALSE | TRUE | 2.6 | 1.336 | 0.344 |
| EHS5-R15 | T7 DP SC RH1 Txn+ | TRUE | FALSE | FALSE | FALSE | TRUE | TRUE | TRUE | -4.6 | -0.482 | -0.124 |
Sample barcoded primer assignment #
| Sample Name | Formal name | Forward Primer Name | Oligo ID | Barcode |
|---|---|---|---|---|
| T7 SC Txn - | EHS0-R0 | PacBioBarFwd-0 | oEH30 | CGGTTAGA |
| T7 SC Txn + | EHS0-R1 | PacBioBarFwd-1 | oEH31 | CCCTCTTT |
| T7 Lin. Txn+ 1 | EHS0-R6 | PacBioBarFwd-2 | oEH32 | CGTTTCTG |
| T7 DP SC Txn+ | EHS0-R8 | PacBioBarFwd-3 | oEH33 | ATGGATCG |
| T7 DP Lin. Txn+ 2 | EHS4-R14 | PacBioBarFwd-4 | oEH34 | GGTAACAC |
| T7 DP SC RH1 Txn+ | EHS5-R15 | PacBioBarFwd-5 | oEH35 | CAAGCAGA |
PCR reaction details #
| Total Volume | 5x Phusion GC buffer | 10 mM dNPTS | 10 uM Primer Mix | Bisulfite converted template | PhuU Polymerase | npH20 |
|---|---|---|---|---|---|---|
| 30 | 6 | 0.4 | 1.5 | 4 | 0.3 | 18.1 |
Reactions were assembled from master mix and then primers and template were added to each reaction aqliuote.
All samples were run for 15 PCR cycles. Annealed at 58.9C for 30 seconds with 2:30 extension time at 72C. Samples then held at 4C for ~2 hours.
Agarose gel extraction PCR products #
Imaged using phone camera on blue light transilluminator.
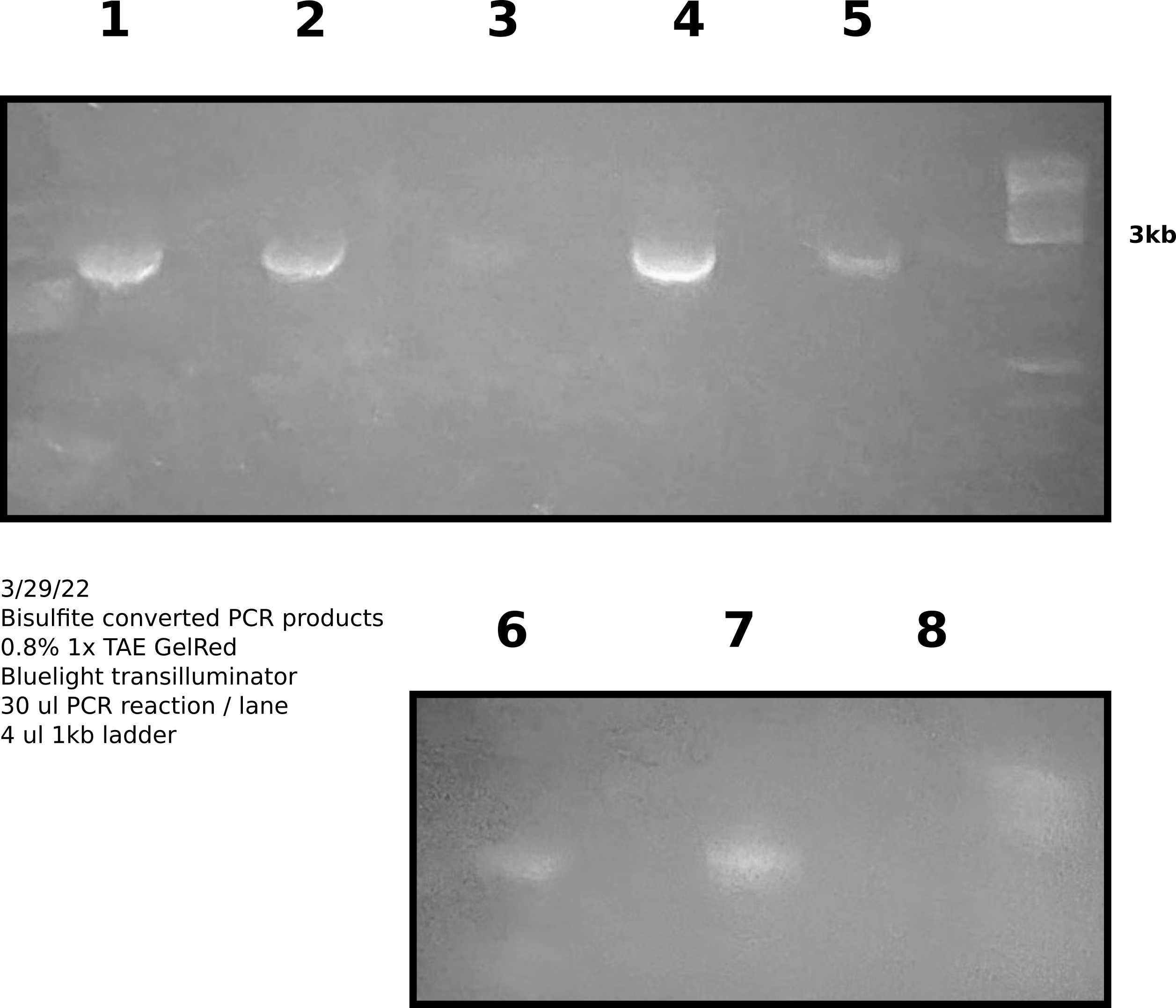
Preformed gel extraction according to Zymogen protocol. Elluted extracted products in 30 ul ellution buffer (10 mM Tris HCl).
Nanodrop extracted products #
Agarose gel extracted products #
| Sample | ng_ul | 260_280 | 260_230 |
|---|---|---|---|
| 1 | 12.4 | 2.299 | 0.019 |
| 2 | 9 | 2.26 | 0.022 |
| 3 | 5.5 | 2.767 | 0.011 |
| 4 | 8.9 | 2.291 | 0.015 |
| 5 | 7.8 | 2.375 | 0.011 |
| 6 | 18.8 | 1.822 | 0.047 |
Testing chloroquine gel protocol using T7Init Mix 0 +- Topo treatment #
Agarose gel image #
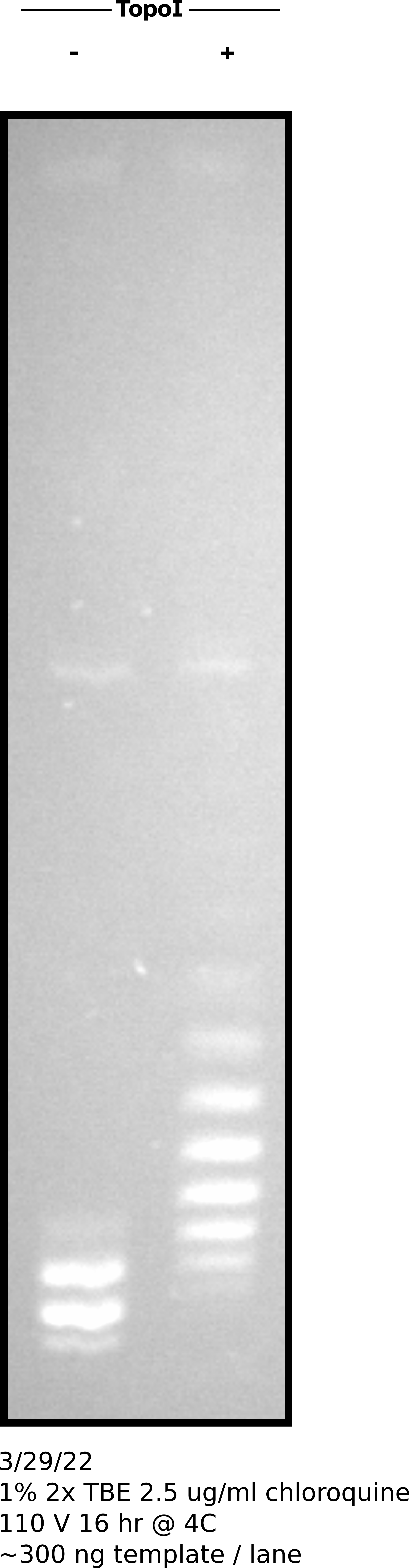
3/30/22 #
Repeat PCR for agarose gel extraction #
PCR reaction was done with same protocol preformed previously with these samples. Ran PCR overnight to run on gel in the morning.
3/31/22 #
Agarose gel extraction of 3/30 PCR products #
Ran complete 30 ul reaction of each sample on agarose gel (1x TAE 60V 2 hrs) and viewed on the trans illuminator (no gel image). However, even though the postive control (using T7InitMix 1 unconverted template) showed a band all bisulfite treated samples except the un-transcribed control did not show a band. Not sure what happened here but seems to be something specific to transcribed reactions and not the PCR master mix. Unfortunately this replicate ends here because of this failed PCR.