QuBit sample measurement #
3/26/22 #
In order to be sure about sample concentrations I re-measured all T7Init Series constructs using the QuBit. I first diluted samples 1:400 (1 ul of sample in 399 ul TE). Then pipetted 2 ul of this dilution into 198 ul of 1x dsDNA QuBit dye. I then allowed samples to warm to room temperature for 5 minutes before measuring samples.
Sample measurements and concentration calculations #
| Insert | Dilution total volume | Measure 1 | Measure 2 | Measure 3 | Stock concentration |
|---|---|---|---|---|---|
| 1 | 400 | 0.534 | 0.52 | 0.513 | 208.9333333 |
| 2 | 400 | 0.622 | 0.606 | 0.603 | 244.1333333 |
| 3 | 400 | 0.434 | 0.426 | 0.42 | 170.6666667 |
| 4 | 400 | 0.283 | 0.268 | 0.268 | 109.2 |
| 5 | 400 | 3.6 | 3.56 | 3.54 | 1426.666667 |
| 6 | 400 | 0.937 | 374.8 | ||
| 7 | 400 | 4.03 | 1612 | ||
| 8 | 400 | 1.41 | 564 | ||
| 9 | 400 | 4.47 | 1788 | ||
| 10 | 400 | 4.68 | 1872 | ||
| 11 | 400 | 4.87 | 1948 | ||
| 12 | 400 | 6.96 | 6.9 | 6.87 | 2764 |
| 13 | 400 | 1.44 | 1.43 | 1.41 | 570.6666667 |
| 14 | 400 | 5.21 | 5.18 | 5.15 | 2072 |
| 15 | 400 | 2.03 | 2.02 | 2.01 | 808 |
| 16 | 400 | 0.654 | 0.649 | 0.646 | 259.8666667 |
| 17 | 400 | 4.74 | 4.72 | 4.7 | 1888 |
| 18 | 400 | 5.1 | 5.07 | 5.05 | 2029.333333 |
| 19 | 400 | 1.1 | 1.1 | 1.1 | 440 |
| 20 | 400 | 5.51 | 5.5 | 5.48 | 2198.666667 |
| 21 | 400 | 8.59 | 8.57 | 8.51 | 3422.666667 |
| 22 | 400 | 11.3 | 11 | 10.9 | 4426.666667 |
| 23 | 400 | 35.1 | 34.8 | 34.5 | 13920 |
| 24 | 400 | 16 | 16 | 15.8 | 6373.333333 |
| 25 | 400 | 4.21 | 4.18 | 4.19 | 1677.333333 |
| 26 | 400 | 2.4 | 2.38 | 2.37 | 953.3333333 |
| 27 | 400 | 2.24 | 2.23 | 2.23 | 893.3333333 |
| 28 | 400 | 7 | 6.98 | 6.97 | 2793.333333 |
| 29 | 400 | 3.83 | 3.8 | 3.76 | 1518.666667 |
| 30 | 400 | 6.01 | 5.99 | 5.97 | 2396 |
| 31 | 400 | 15.6 | 15.6 | 15.5 | 6226.666667 |
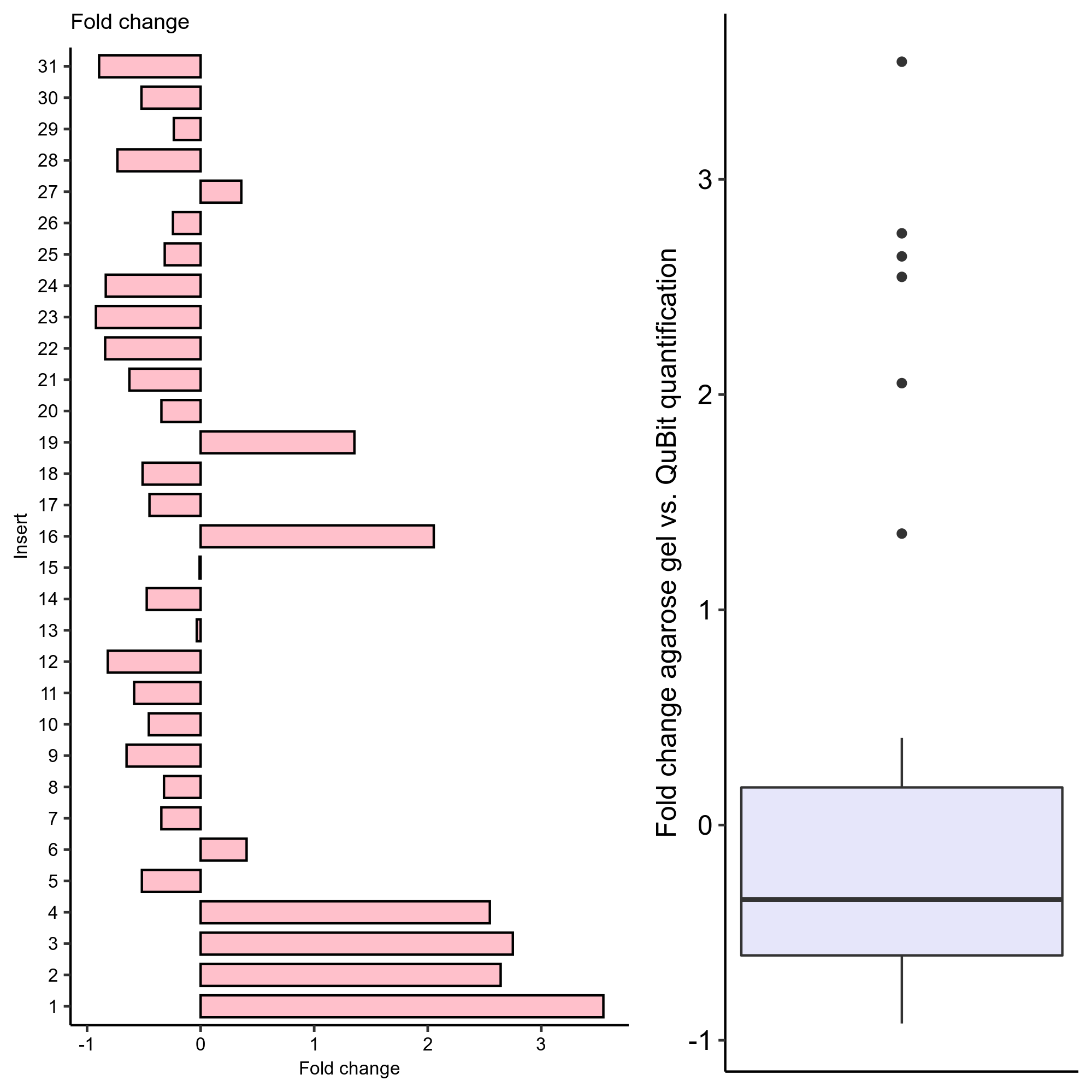
Concentrations were very different than previously measured and did not make much sense with Qubit claiming some samples are more than 20X more concentrated than others which seems highly unlikely.
To follow this up I ran a gel using 1 ul of each construct as well as pFC9 and pFC53 which are labeled at 300 and 500 ng/ul respectively as loading controls.
Agarose gel follow-up to wacky QuBit results #
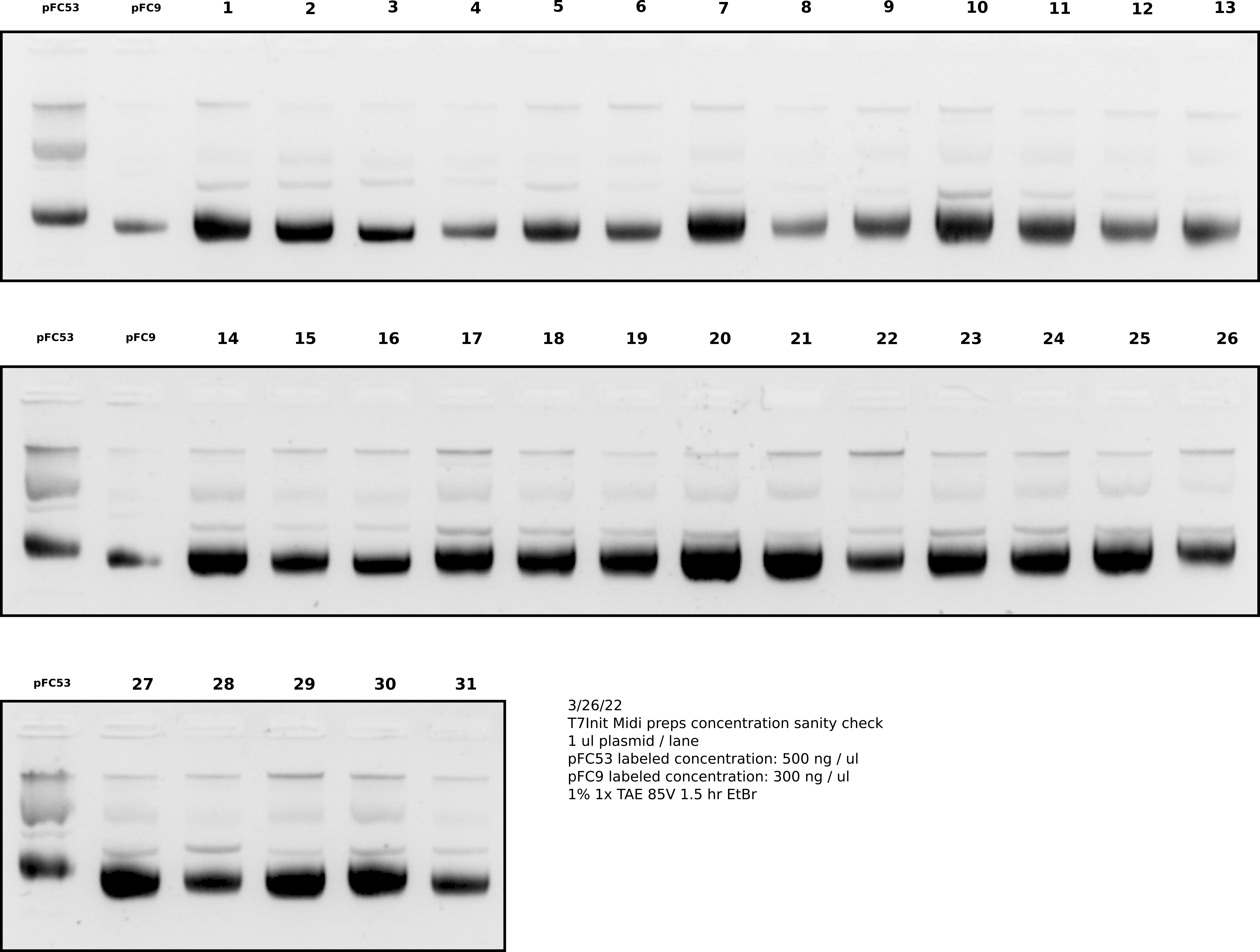
Analysis: By eye #
By eye it is clear the relative concentrations between constructs measured by the QuBit are way off. Quantification by gel may be the most reliable for creating a plasmid mix. While exact mass may be a bit off the relative concentrations are likely to be more accurate which is arguably more important at the sequencer to avoid an extreme over-abundance of one construct over another.
Analysis: Quantification using ImageLab #
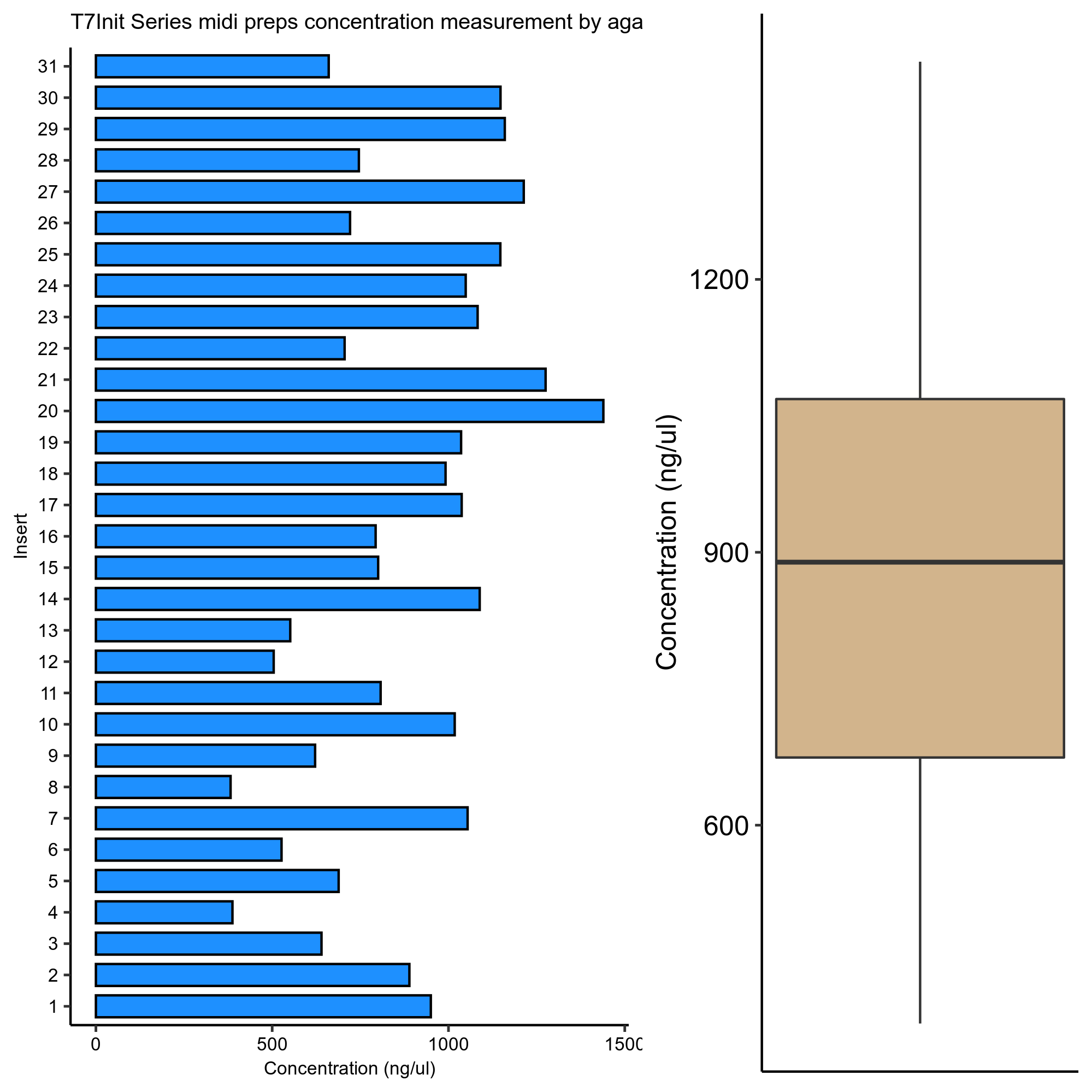
I then quantified the gel using the BioRad ImageLab software. Plots and all calculations were made in this Jupyter notebook.
Concentration measurements are shown in the table below. These measurements
informed the volumes of each construct in T7InitMix-1 which was used
as the primary substrate for all samples.
Sample concentration measurements by agarose gel
| Band_Label | ul_add_to_mix | insert_num | ng/ul |
|---|---|---|---|
| VR1 | 3.15808 | 1 | 949.945 |
| VR2 | 3.37399 | 2 | 889.155 |
| VR3 | 4.68847 | 3 | 639.868 |
| VR4 | 7.74572 | 4 | 387.311 |
| VR5 | 4.35732 | 5 | 688.496 |
| VR6 | 5.69779 | 6 | 526.52 |
| VR7 | 2.84563 | 7 | 1054.25 |
| VR8 | 7.85345 | 8 | 381.998 |
| VR9 | 4.82521 | 9 | 621.734 |
| VR10 | 2.9479 | 10 | 1017.67 |
| VR11 | 3.71493 | 11 | 807.552 |
| VR12 | 5.94932 | 12 | 504.259 |
| VR13 | 5.44227 | 13 | 551.24 |
| VR14 | 2.75593 | 14 | 1088.56 |
| VR15 | 3.74733 | 15 | 800.571 |
| VR16 | 3.78123 | 16 | 793.392 |
| VR17 | 2.89164 | 17 | 1037.47 |
| VR18 | 3.02504 | 18 | 991.723 |
| VR19 | 2.89629 | 19 | 1035.81 |
| VR20 | 2.08432 | 20 | 1439.32 |
| VR21 | 2.35206 | 21 | 1275.48 |
| VR22 | 4.25264 | 22 | 705.444 |
| VR23 | 2.77091 | 23 | 1082.68 |
| VR24 | 2.86016 | 24 | 1048.89 |
| VR25 | 2.61526 | 25 | 1147.11 |
| VR26 | 4.16159 | 26 | 720.878 |
| VR27 | 2.47214 | 27 | 1213.52 |
| VR28 | 4.02208 | 28 | 745.883 |
| VR29 | 2.5868 | 29 | 1159.74 |
| VR30 | 2.61421 | 30 | 1147.57 |
| VR31 | 4.54334 | 31 | 660.307 |
Absolute quantification’s #
Fold change to QuBit concentration measurements #