Background #
The T7Init series of plasmids is transcribed using the T7 phage polymerase. For future IVT experiments that will be subjected to SMRF-seq it will be important to have a bases for the ratio of polymerase to DNA used in a given reaction.
Given that we know transcription of pFC9 results in >95% R-loop formation observed by agarose gel, I plan to determine the minimal T7 to DNA ratio that will produce this degree of shift in pFC9 and use this as the T7 to DNA ratio for all future T7 IVT experiments.
Optimizing NEB T7 for IVT #
3/4/22 #
Experimental setup #
I am interested in determining the minimal ratio of T7 (in units) to DNA (expressed in nM) that can be used for a complete pFC9 IVT reaction. I designed IVT reactions with decreasing concentrations of NEB T7 and constant DNA concentration (5nM) which was informed by DNA concentrations used in the lab IVT protocol and NEB provided protocol for T7 transcription reactions.
The complete table describing reaction setup, dilutions and reagent lot numbers is available at this link. The reactions are summarized in the table below.
| Sample Number | Volume Transcription master mix | Polymerase Dilution | Polymerase Dilution Volume | npH20 | Pol units / nM Template | Template concentration (nM) | Polymerase concentration (units) |
|---|---|---|---|---|---|---|---|
| CTRL | 8 | CTRL | 0 | 2 | 0 | 5 | 0 |
| STOCK | 8 | STOCK | 2 | 0 | 20 | 5 | 100 |
| 1 | 8 | 1 | 2 | 0 | 10 | 5 | 50 |
| 2 | 8 | 2 | 2 | 0 | 5 | 5 | 25 |
| 3 | 8 | 3 | 2 | 0 | 2.5 | 5 | 12.5 |
| 4 | 8 | 4 | 2 | 0 | 1.25 | 5 | 6.25 |
Experimental protocol #
Prepared master mix as described by experiment table. Add respective T7 dilutions to each sample and incubated at 37C for 20 minutes in the thermocycler. After incubation stopped transcription reaction by adding EDTA to final concentration of 36 mM (6x transcription buffer Mg concentration).
Digested sample with 1 ul of 10 mg / ml RnaseA for 15 minutes at room temperature and then ran complete sample on an agarose gel equating to ~230 ng of DNA per lane.
Agarose gel image #
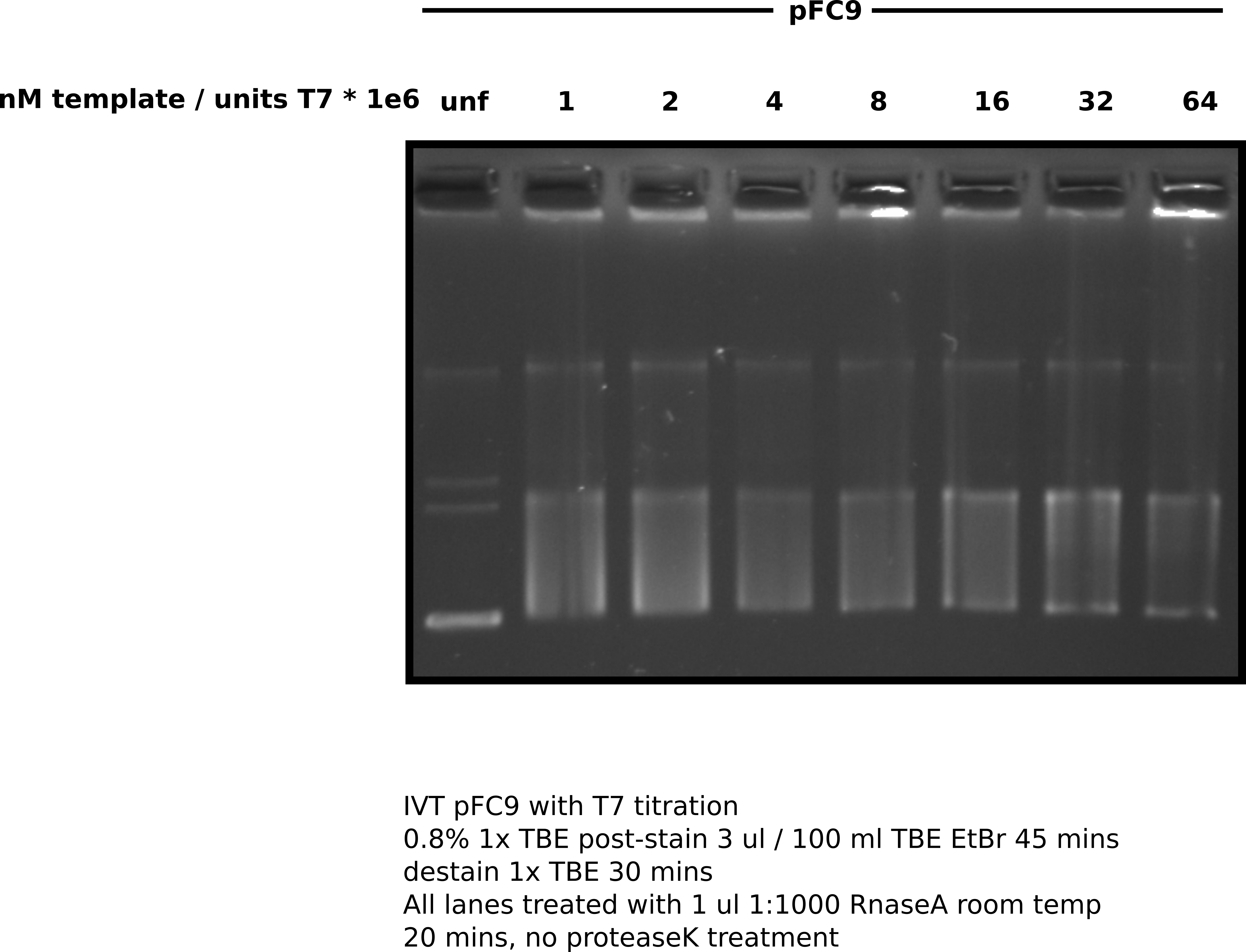
A note on the units used in this gel
This gel quantifies polymerase concentrations as the number of nanomoles of template / units of polymerase present in the transcription reaction times 1e6. Nanomoles of template / units pol is generally a very small number so multiplying by 1e6 just makes it more human readable.
Analysis #
Based on the results of the above gel it appears that reducing T7 concentrations relative to DNA template does not result to sharp decreases in R-loop formation indicated by band shift and RnaseA insensitive smear compared to the un-transcribed control. 3 is about what the current lab protocol calls for so I will continue with that ratio for future transcription experiments.