Background #
With the latest chemistry and sequencer, PacBio can achive up to 4 million reads per SMRT-cell. Even at 60% of this, this allows for thousands of reads per insert per sample; even if each sample contains a transcribed, RnaseH treated and un-transcribed control. This means we could easily jam T7Init series, TacTermination series and Cas9 treated samples onto one SMRT-cell as long as there is some way to tell treatments apart.
Since for first round of sequencing I am planning on just having the genome center prepare libraries it would be best to send as few samples as possible since they charge by the sample. So ideally we get all treatments into the same tube with a way to tell them apart computationally.
Using overhanging barcoded PCR primers #
One approach is to use overhanging PCR primers with unique barcodes (per sample) during the PCR reaction required after bisulfite conversion of plasmid sequences. Each treatment would be amplified at the same target site but have a unique overhanging sequence that could be later picked out during data analysis.
Barcoded overhanging primer design #
I designed primers for this application in this jupyter notebook. Each primer includes a 10 bp buffer region at the 5' end followed by a unique 8 bp barcode. The buffer serves to protect the barcode from anything that might damage 5' ends of sequences during library prep. I used DNAchisel to ensure barcodes and buffers did not contain EcoRI, SacI, KpnI or HindIII sites and avoided A homopolymers and direct repeats while maintaining between 0.4 and 0.6 GC content and maximizing hamming distance between other barcodes to ensure each is easily identifiable.
T7Init series example #
Below is a simulated PCR reaction on pFC9VR1 using one of the barcoded forward primers and the T7Init reverse primer.
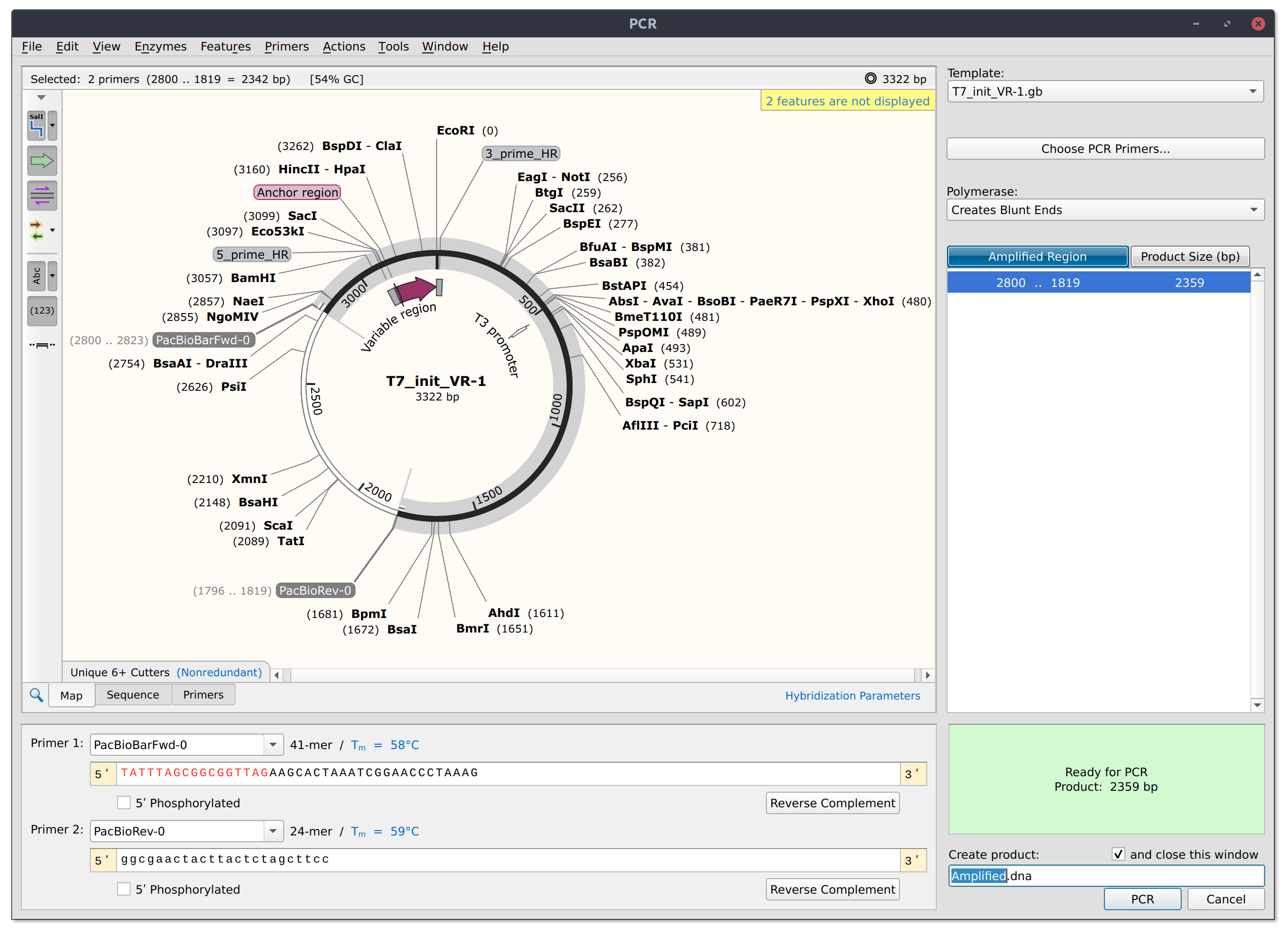
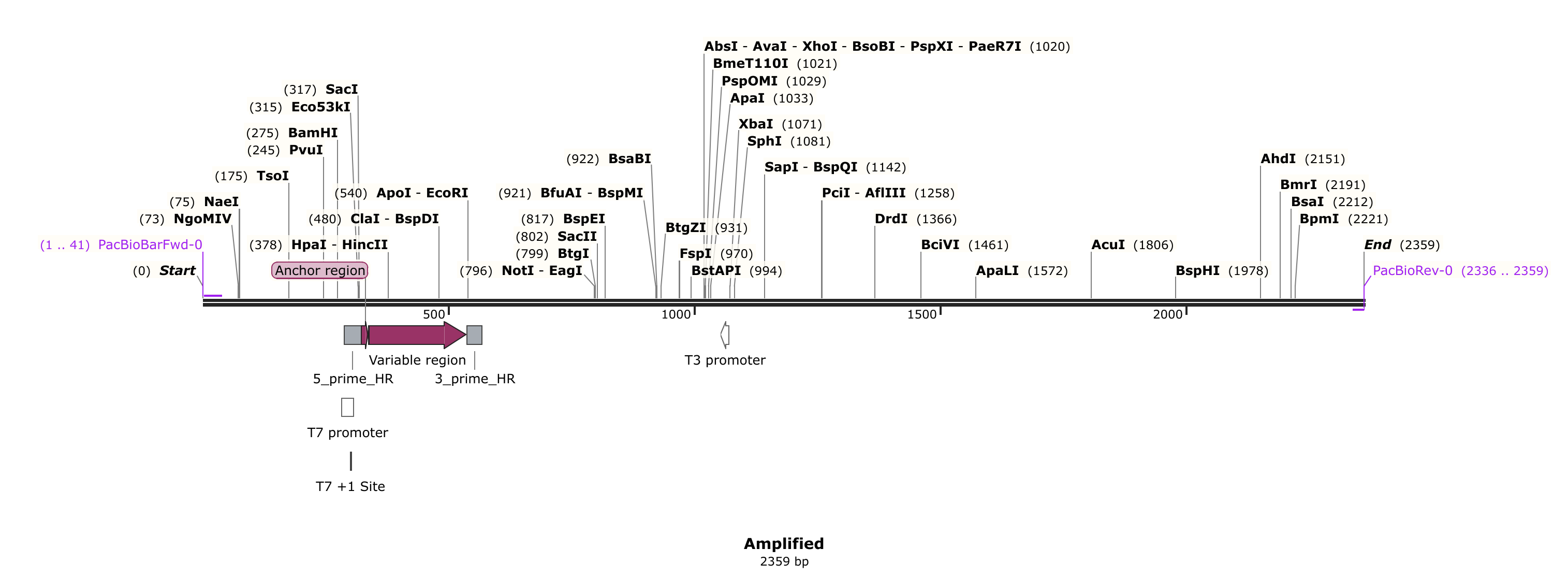
Linear depiction of the product is shown below. After library prep this product would be subjected to PacBio sequencing.
Current oligos and order status are available in the PacBioBarcodedPrimers sheet of the Ethan’s oligos table.