PacBio library prep notes and protocols #
Malika lab notes #
PCB-04 folder #
- Notes on footprinting “Set 2 PLB”.
- Notes for prepping genomic DNA for footprinting.
- PCR amplification of libraries using PhusionU pol with GC buffer for 30 cycles.
- Includes PacBio 2kb Template Prep and Sequencing protocol but does not mention
use of PacBio Template prep kit.
- Assuming this kit is used though because no mention on making homemade reagents.
- PCR follows bisulfite conversion reactions. After PCR preforms 1x Ampure bead wash.
In vitro footprinting folder #
This seems like what I am looking for. PacBio library prep notes related to pFC53 footprinting data.
- Inculded PacBio 2kb template prep protocol with annotations
Genome center sample requirements #
Sample quantity #
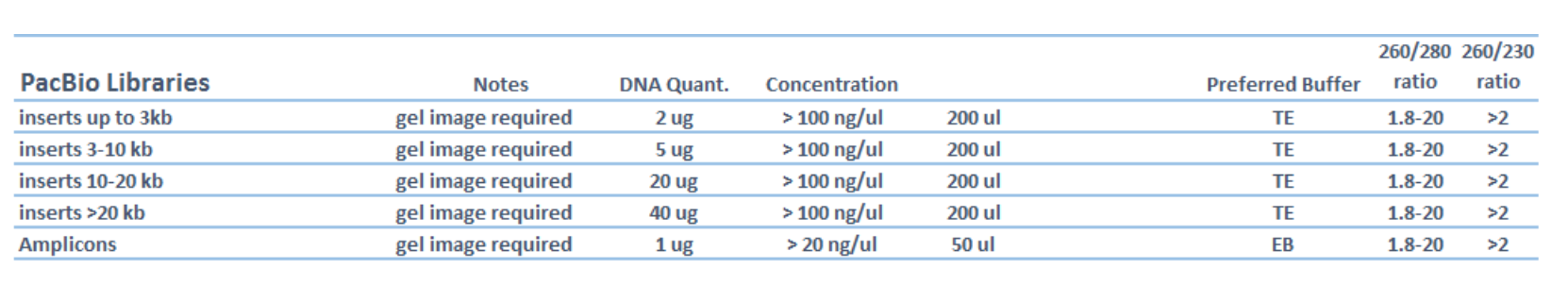
Fragments will be around 3 kb so should include 5 ug of sample if delivered
for library prep. For 33 plasmids (31 VR inserts, positive and negative controls)
that is 0.161 ug or 161 ng of each plasmid. NEB recommends up to
1 ug DNA
template per 50 ul T7 reaction.
So when completing actual protocol would use 1 ug total of all plasmids then split into 2 500 ng samples for bisulfite conversion as recommended DNA mass for the kit is 500 ng. After bisulfite conversion re-measure DNA concentration and PCR. Will need to determine the number of cycles in order to amplify to sufficient concentration. To me it would make sense to PCR a large amount of DNA in order to minimize duplicates.