12/8/21 Meeting #
Playing with plasmid length #
- Natasha brought up plasmid length on R-loop formation patterns. Fred
suggested as side project if possible.
- Would be easiest to decrease size. Find double cutters with significant distance between them or other distance compatible restriction sites. However, would also need to avoid removing amp resistance gene and the ORI. This might make extending plasmid length easier.
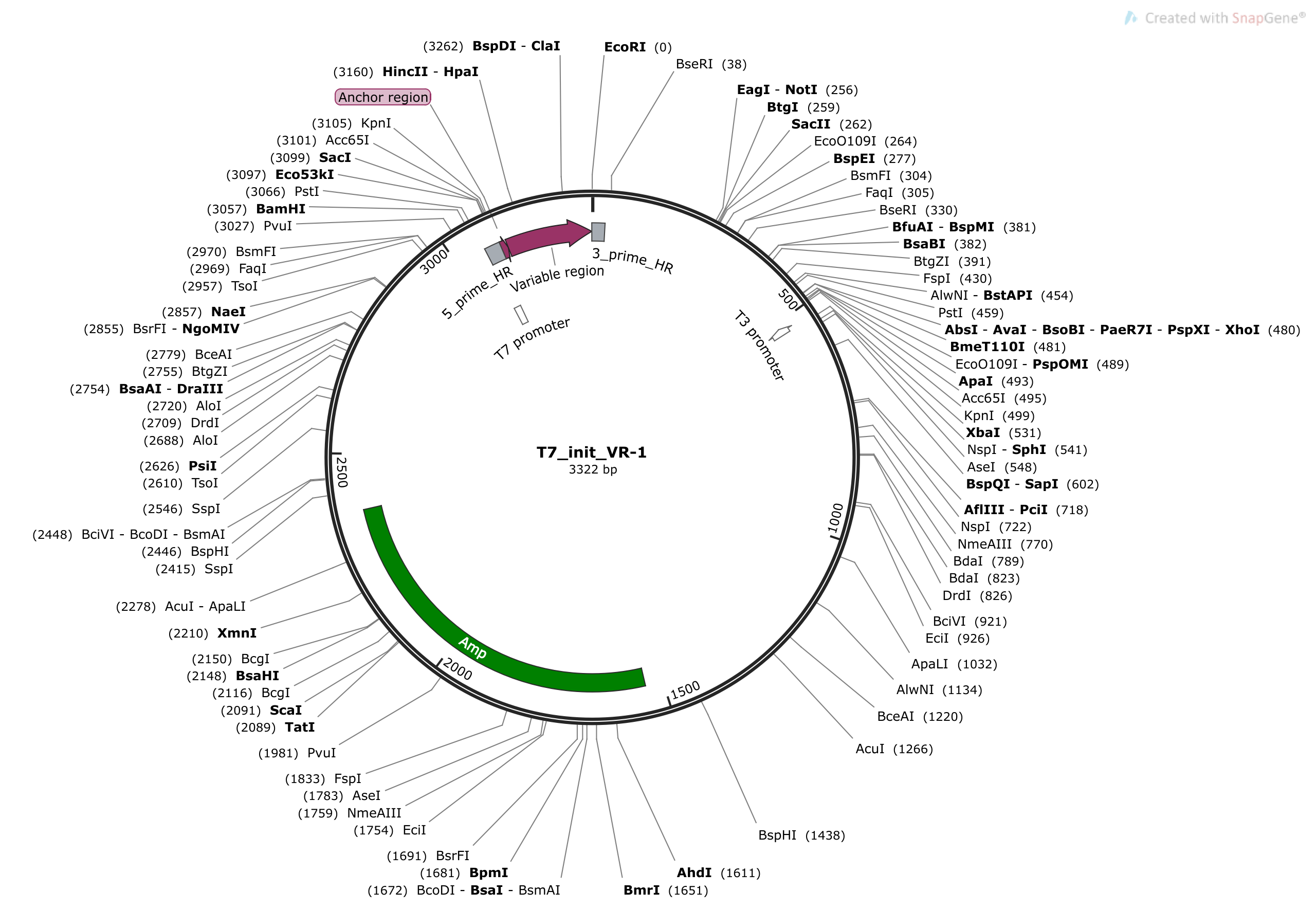
There is actually a relatively small window that plasmid could be cut within. It would have to be between the Amp gene and start of the T7 promoter in order to avoid R-loop forming region and the ORI.
One approach could be to use two double cutters to produce compatible fragments. Agarose gel extract one of the fragments and then digest again with the fragment you want to add in to extend the length. Image below is example using DrdI and PstI.
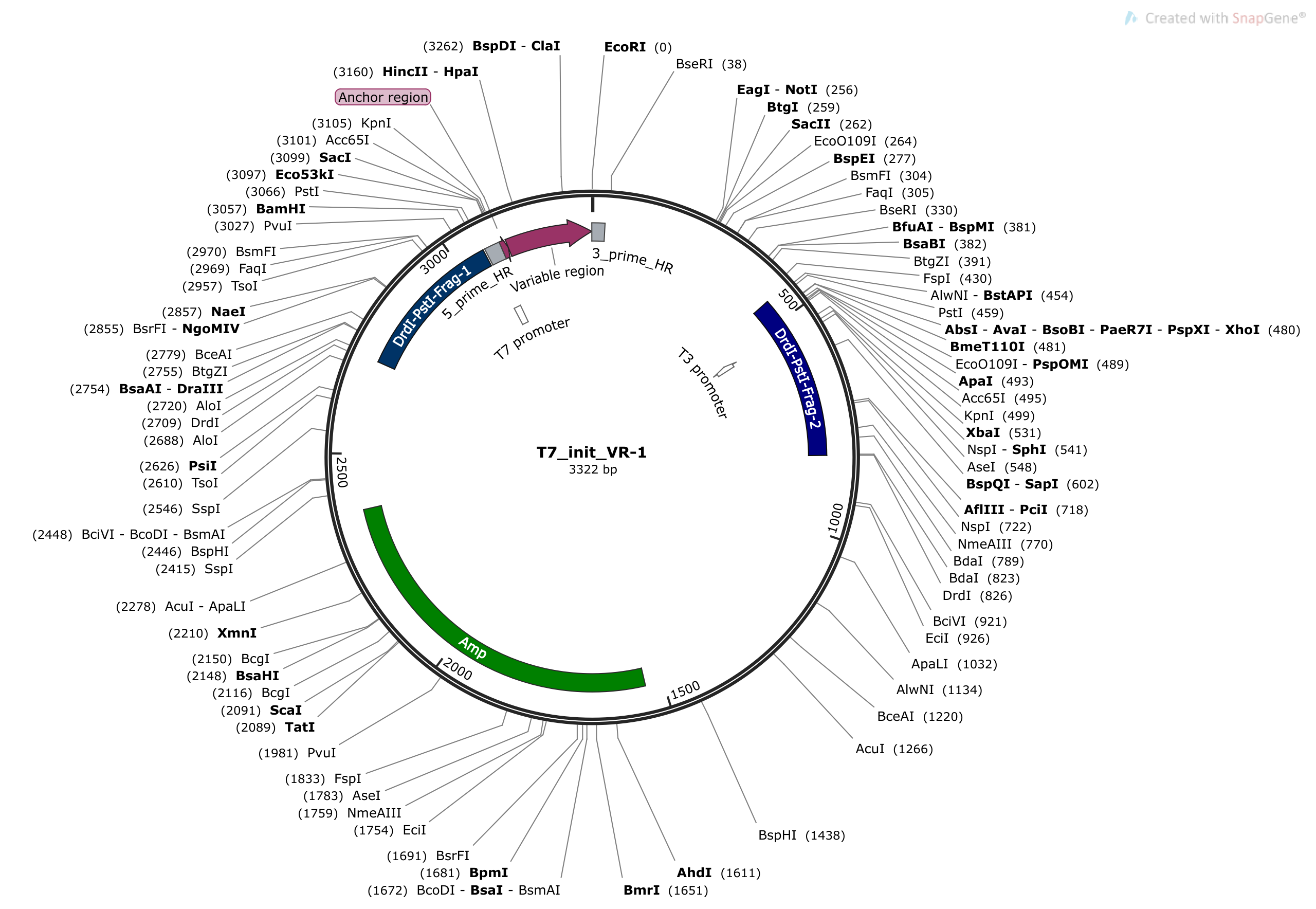
This idea will not work! (probably)
This will not work because when digesting with the same enzymes (which are both double cutters) the plasmid will be cut at four locations preventing predictable ligation patterns so fragments A and B (for example) may end up in opposite locations. This would also require two ligation events instead of 1 to reconstitute the intact plasmid.