Nick validation #
The interpretablity of these experiments hinge on being able to place a nick very precisely within a plasmid. Therefore it is nessicary to have a method to validate the nicks made with the Cas9 system. After talking with Fred about this and bouncing some ideas back and forth I suggested that it might be possible to validate nicks by Sanger sequencing. My thinking is that since Sanger sequencing only deals with one strand at a time, if you sequence the strand the nick is present on the read should die at the site of the nick and continue normally if no nick is present. If there is some mixture of nicked and un-nicked template then that should be reflected by a drop in signal in the trace.
1/18/22 #
Testing Sanger sequencing as a method for nick validation: Experimental design #
I’ve already bought the nicking endonuclease, Nt.BsQI and validated its activity. Nt.BsQI can be used to simulate and cas9 induced nick on a plasmid.
Template DNA and sequencing primer #
pFC8tacT1T2 has two Nt.BsQI sites, one of which is nearby the Tac promoter, shown in the plasmid map below.
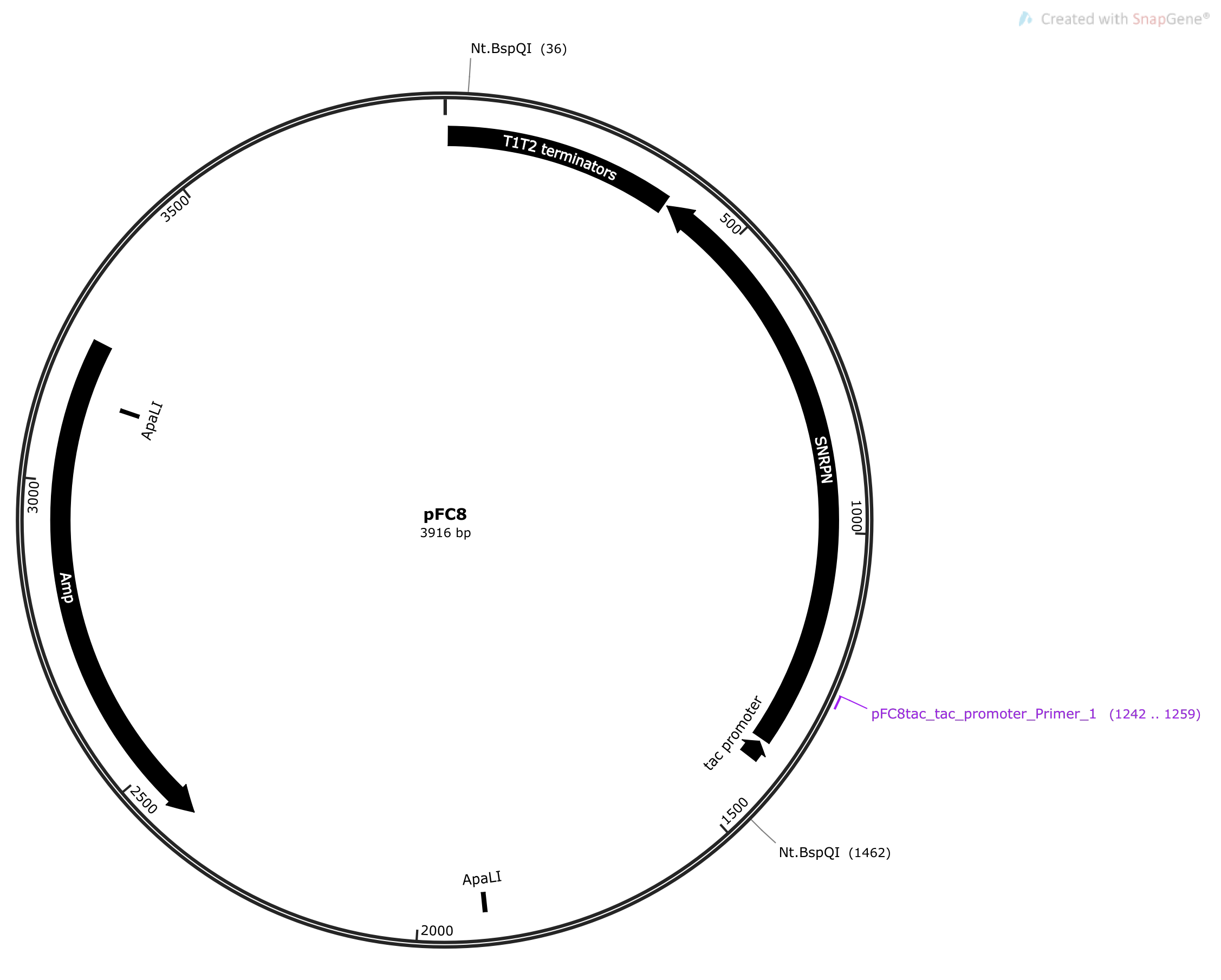
This is also near and on the opposite strand
as the sequence for pFC8Ttac_tac_promoter_Primer_2. This means that
pFC8Ttac_tac_promoter_Primer_2 will bind to the nicked strand and can be used
for sequencing. The image below shows both pFC8Ttac_tac_promoter_Primer_1
and pFC8Ttac_tac_promoter_Primer_2 on pFC8tacT1T2 template and the nick
induced by treatment with Nt.BspQI.
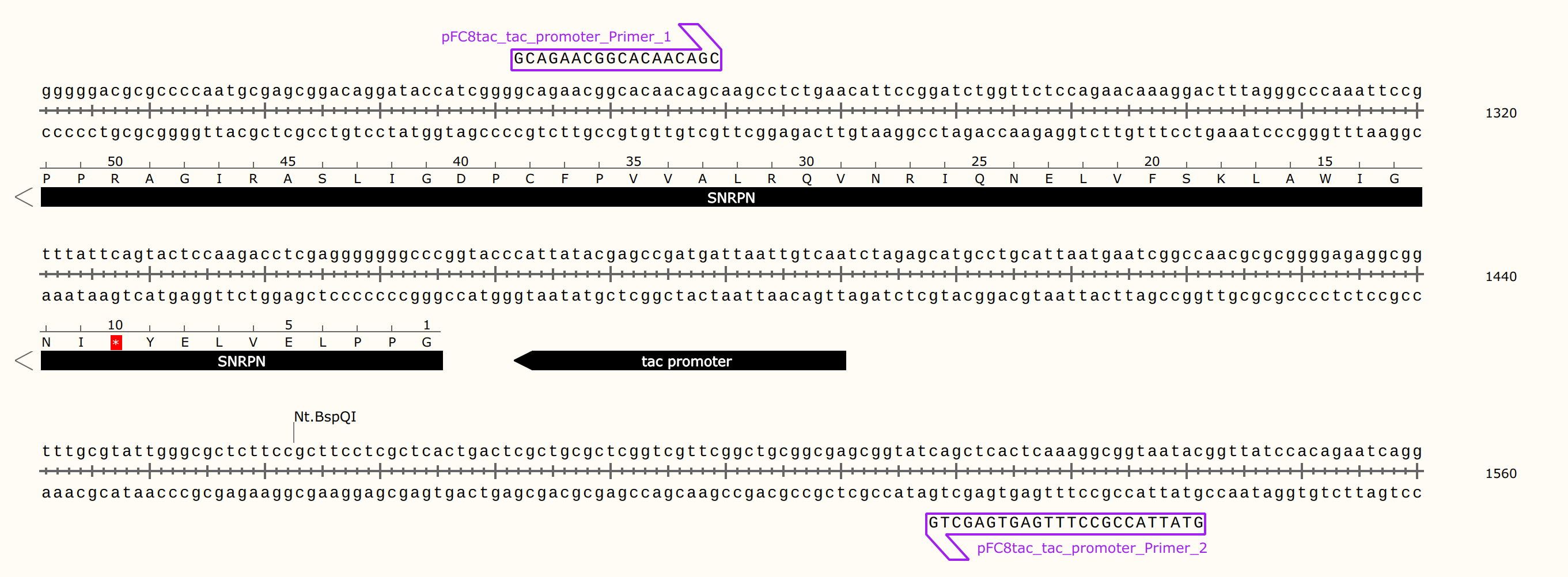
The forward strand will be nicked. So the primer must be complementary to this strand so it will act as the template for the polymerase during the Sanger reaction.
1/19/22: Correction to validation protocol
I previously specified that
pFC8Ttac_tac_promoter_Primer_1should be used for this validation experiment, this was incorrect as this primer will bind to the reverse strand resulting in use of the reverse strand as the template. Since the nick is on the forward strand we can expect sequencing to be unaffected by the nick. This is now corrected topFC8Ttac_tac_promoter_Primer_2.
Protocol #
| Sample | Nt.BspQI (ul) | pFC8tacT1T2 (ul) | R3.1 10x Buffer | npH20 | Total Rxn Volume |
|---|---|---|---|---|---|
| 1 | 1 | 7 | 2 | 10 | 20 |
| 2 | 0 | 7 | 2 | 11 | 20 |
- Prepare samples as above, add enzyme last.
- Incubate samples at 50C for 60 minutes.
- Heat inactivate at 80C for 20 minutes.
- Run 1 ul of each sample on 0.8% agarose 1x TAE gel 100V for 1 hour.
- Move 12.5 ul of each sample to new PCR tubes.
- Add 2.5 ul of
pFC8tac_tac_promoter_Primer_2to each sample. - Submit for Sanger sequencing through Quintara.
1/22/22 #
Tadas followed revised Nt.BspQI protocol using pFC8TacT1T2 as the template and sent for sequencing. I then aligned reads back to reference to produce the map shown below.
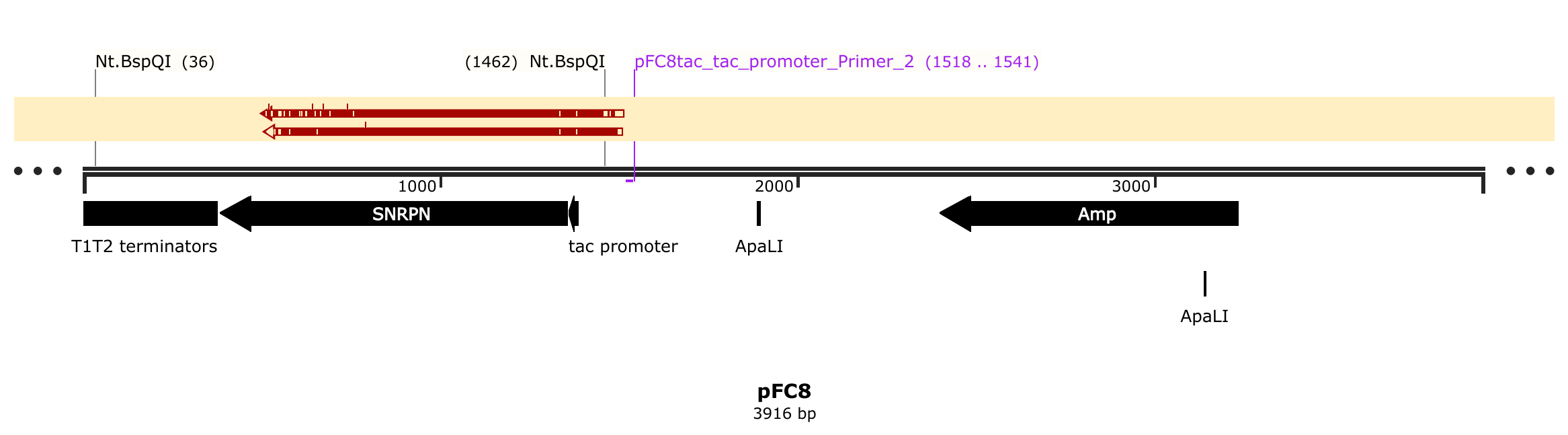
The top read is the nicked sample and bottom is the control. Based on this result alone nicking seems to have basically no effect on the sequence read.
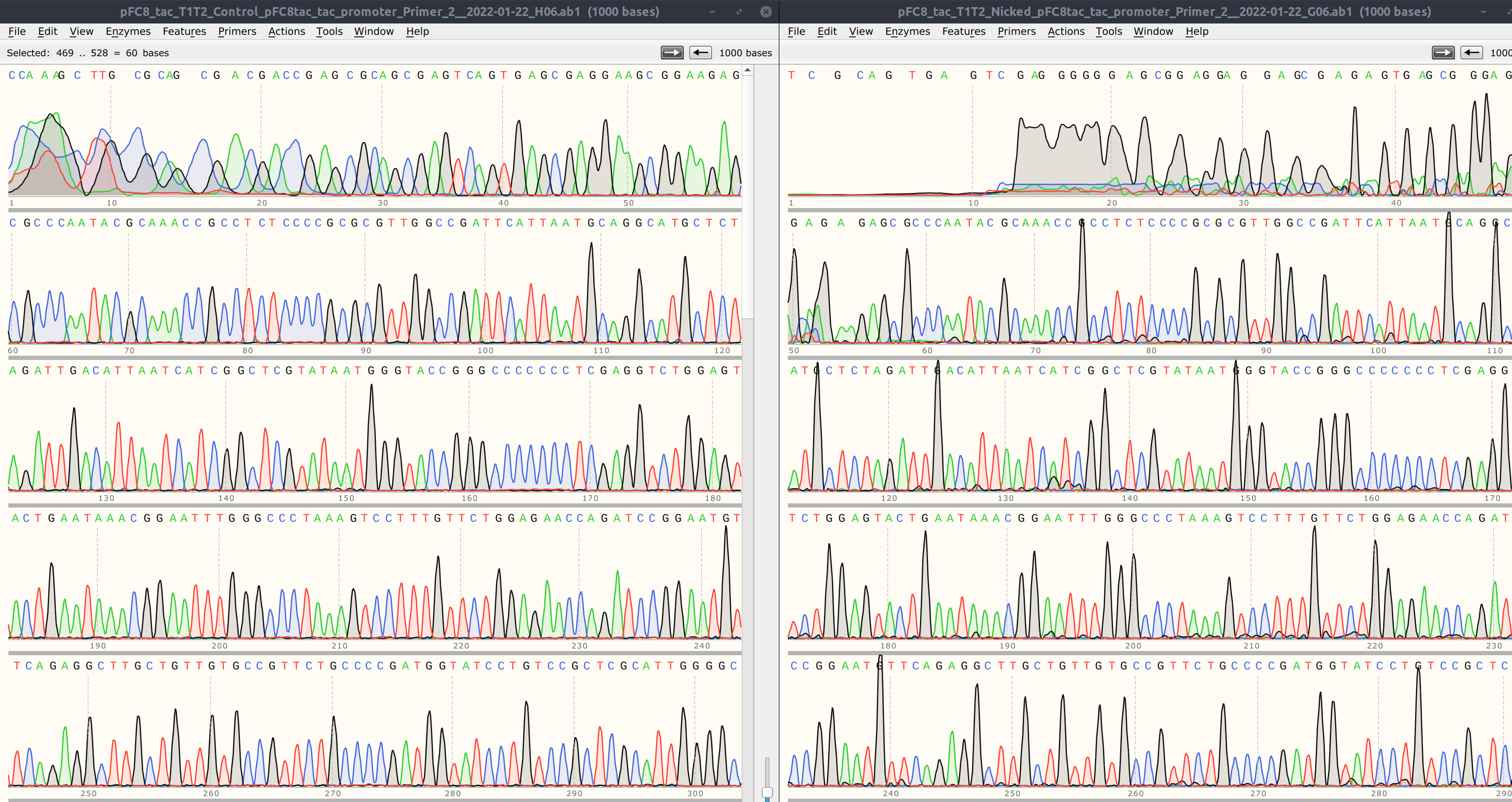
Comparing the traces (shown above, un-nicked on left nicked on right) shows that reads are both strong besides first ~20 base pairs or so which is to be expected. Only major difference is lack of signal for first ~5 bp of the nicked read.
1/24/22 #
Nick validation strategy update #
pFC9 Nt.BspQI nick sanger sequencing through Davis-seq #
Followed nick validation test protocol
but used pFC9 instead of pFC8tacT1T2. pFC9 only has 1 Nt.BspQI recognition
site opposed to pFC8tacT1T2’s 2. Only change I made to the protocol was after
heat killing Nt.BspQI I boiled samples at 100C in thermocycler for 5
minutes and then immediately placed onto ice to prevent re-annealing. The motivation
being that boiling the samples and then rapidly cooling, and keeping them
cold should help ensure that the DNA is single stranded during the sequencing
reaction and to hopefully increase the reaction’s sensitivity to nicks.
Before boiling I took 1.5 ul of each sample (nicked and un-nicked) and ran on
a gel which is shown below in order to confirm that Nt.BspQI treatment
was successful.
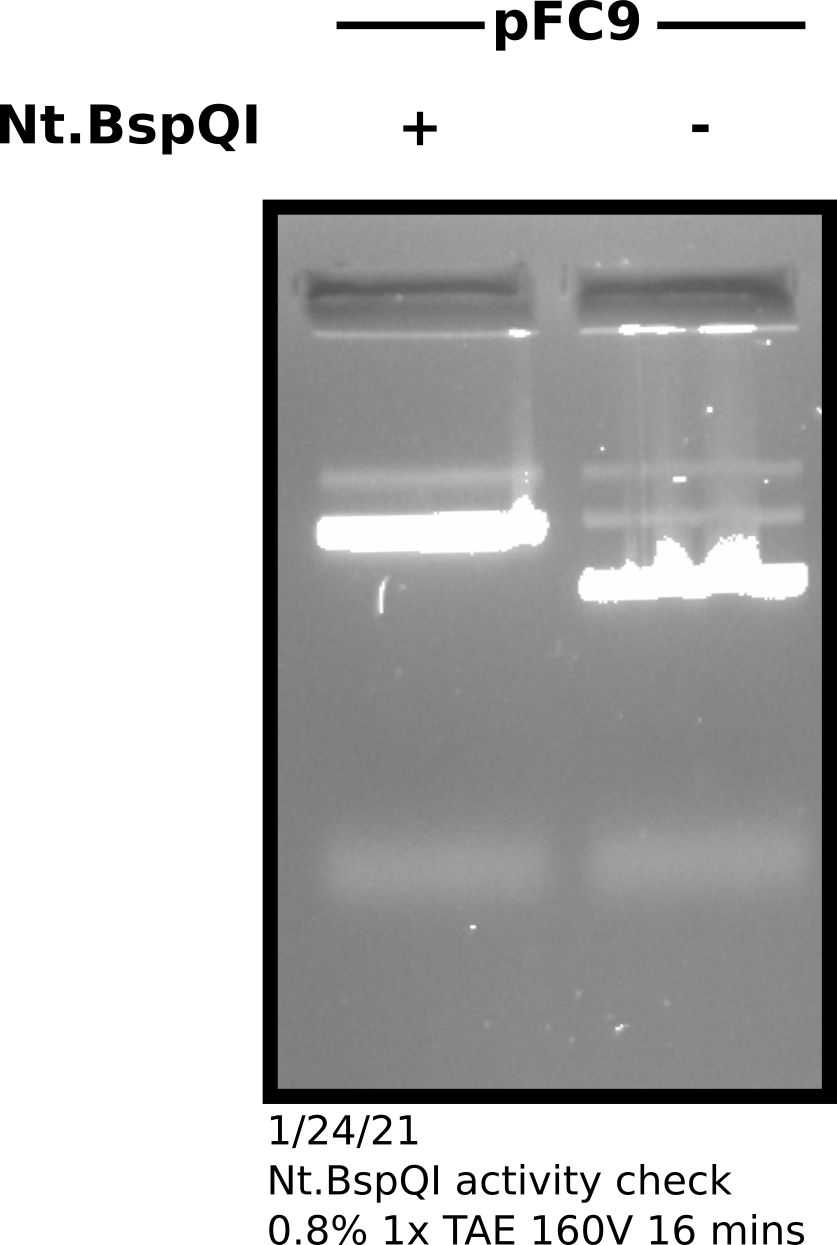
Nt.BspQI successfully nicked by treated sample with close
to 100% efficiency.I brought samples on ice to the UC Davis sanger sequencing center. Did not send to Quintara because I cannot be sure the samples are kept cold by Quintara. Important details from the order form are shown in the table below.
| Name: | Ethan Holleman | Date: | Order number: | 41001 |
|---|---|---|---|---|
| Email: | etholleman@ucdavis.edu | 1/24/2022 | Phone: | 8479225317 |
| PLATE | “DNA SAMPLE NAME” + “PRIMER NAME” | PREP METHOD | PLASMID/PCR SIZE | [DNA] |
| A01 | PFC9 + P2 | None | 3.5 kb | DNA |
| B01 | pFC9-nicked + P2 | None | 3.5 kb | DNA |
Sequencing results #
I received sequencing results 2 days later on the 26th, and the results look good!
The first trace shown below is the un-nicked sample (pFC9-nicked + P2 ). Traces
are also downloadable at this link.
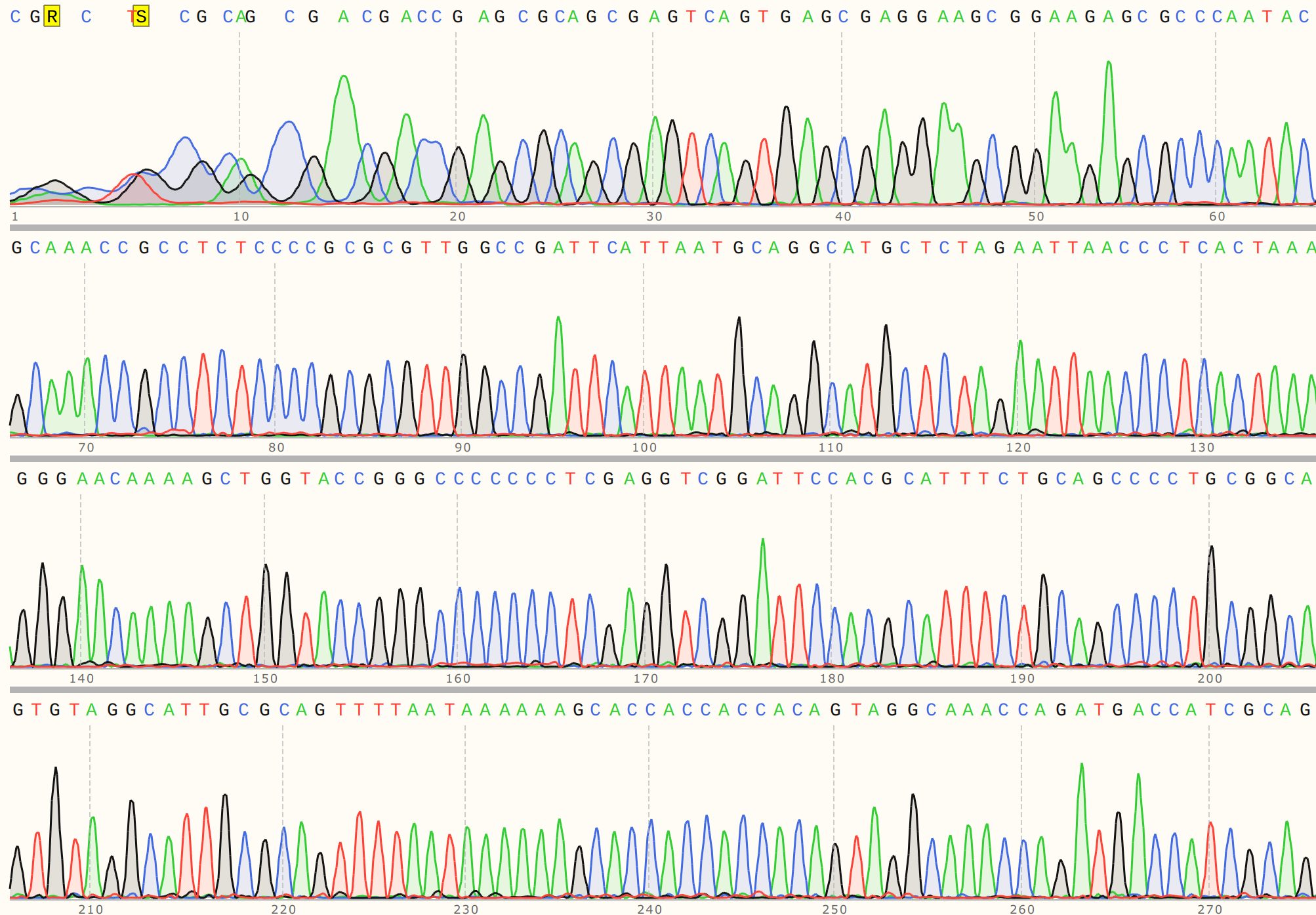
And the nicked sample.
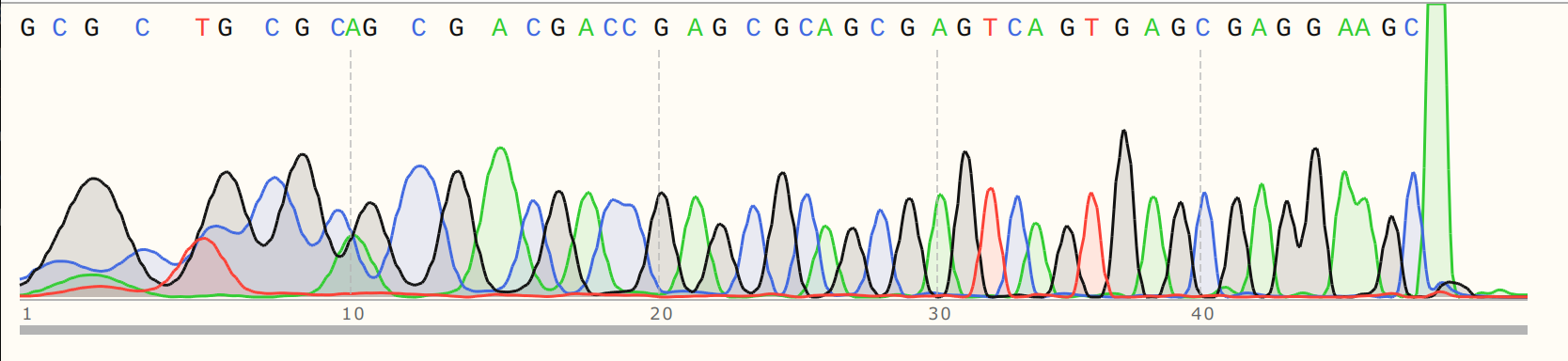
I then aligned these reads to the pFC9 reference using SnapGene and as expected
the nicked read dies exactly at the Nt.BspQI site (the top read is the nicked
sample).
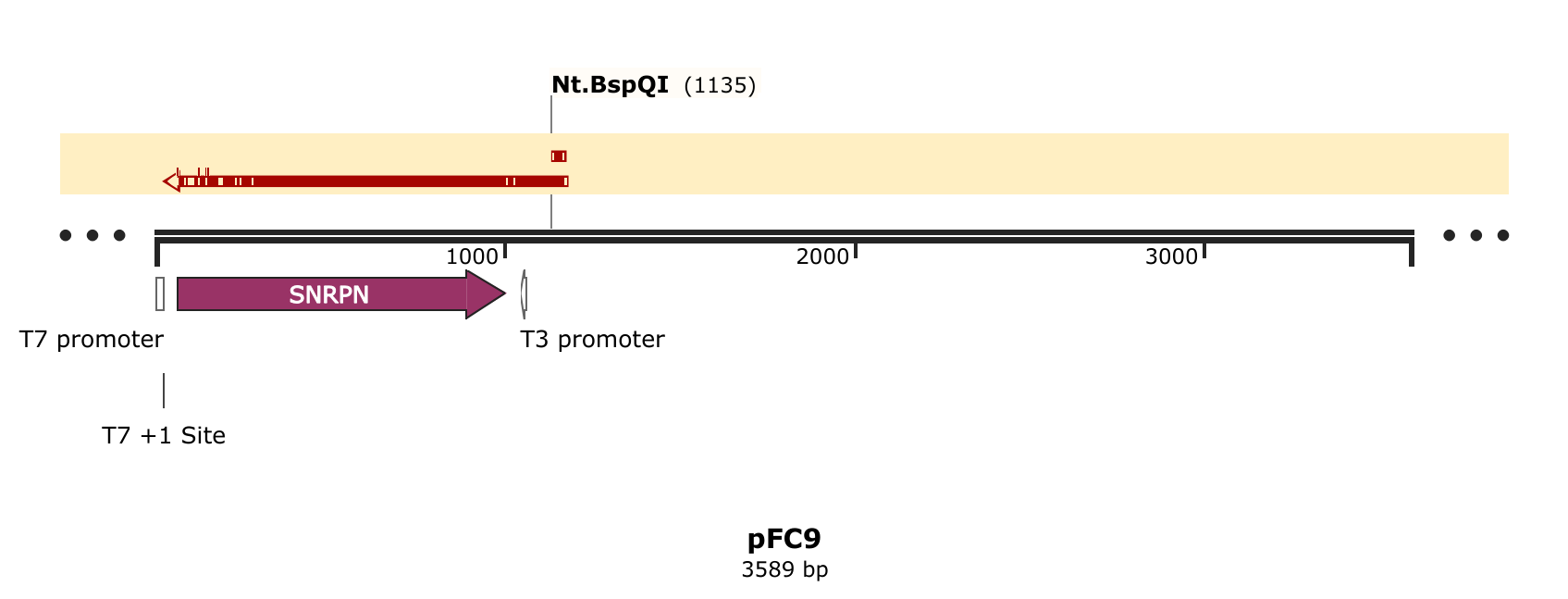
Moving forward will continue to use this protocol and Davis sequencing for nick validation.
Nt.BspQI sanity check #
In order to confirm that this method is robust enough for nick detection with Cas9 (probably should have done this earlier) testing with multiple replicates of Nt.BspQI treated samples.
3/17/22 #
In separate reactions digested 800 ng of pFC9VR18 with 4 ul NEB Nt.BspQI enzyme in total volume of 150 ul using NEB 10x r3.1 buffer. Allowed to digest for 2 hours at 37C. After digestion increased volume to 200 ul and digested sample with BamHI-HF with NEB 10x rCutSmart buffer. Allowed BamHI digestion to proceed for 2 hours at 37C. Then preformed phenol/chloroform precipitation on all samples (8 total, 6 treated with BspQI and 2 untreated). Resuspended all samples in 7 ul Tris-HCl and froze for overnight storage.
3/18/22 #
In the morning thawed all samples and then boiled for 5 minutes in the thermocycler. Immediately after boiling spun down samples and froze by placing in the -80C freezer.
Prepared samples for sequencing at the Davis center. Labels are according to the table below.
| Sample Number | Primer | Primer number | Treatment | DavisSeq Sample name |
|---|---|---|---|---|
| 1 | oEH2 | 2 | Nt.BspQI | 1 + p2 |
| 2 | oEH2 | 2 | Nt.BspQI | 2 + p2 |
| 3 | oEH2 | 2 | Nt.BspQI | 3 + p2 |
| 4 | oEH24 | 24 | Nt.BspQI | 4 + p24 |
| 5 | oEH24 | 24 | Nt.BspQI | 5 + p24 |
| 6 | oEH24 | 24 | Nt.BspQI | 6 + p24 |
| 7 | oEH2 | 2 | Untreated | 7 + p2 |
| 8 | oEH24 | 24 | Untreated | 8 + p24 |
Original order form and data are available on google drive at this link.
3/21/22 #
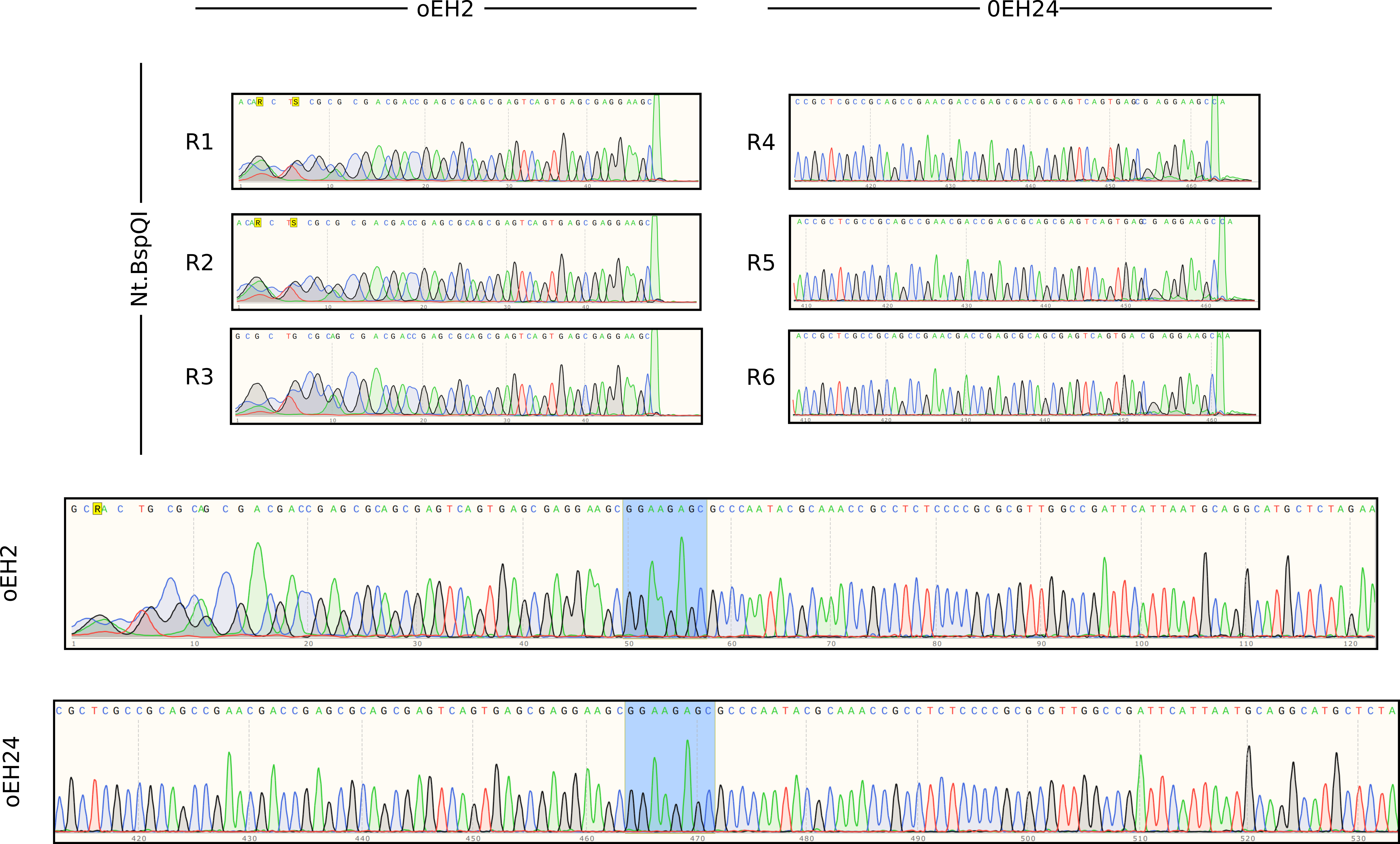
Received reads via email and analyzed results using SnapGene. First looked through all traces which are summarized in the figure below.
Read traces #
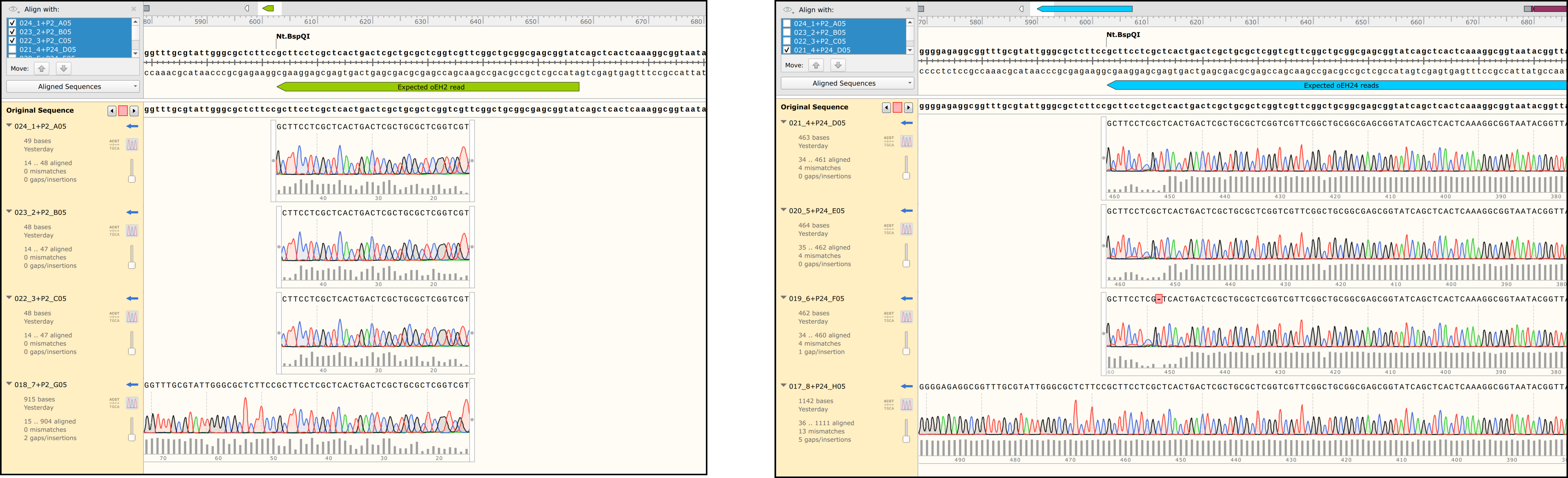
Read alignments by primer used #
Analysis #
Based on these results highly targeted nicks as those that would be expected to be induced by Nt.BspQI are consistently able to terminate Sanger sequencing reads regardless of the distance the sequencing primer is placed away from the nick.
Therefore inconsistencies between high nicking efficiencies as observed by agarose gels and low efficiencies implied by lack of terminated Sanger reads when using Cas9 can most likely be explained by off target nicks.