Hypothesis #
Single strand nicks on the coding strand serve as strong R-loop initiators.
Experimental Design #
To test if nicks can initiate R-loops in vitro I will use a Cas9 nickase to target specific sequences in VR insert plasmids. Using multiple guide RNAs I will attempt to alter the location of initiation in constructs that already show strong R-loop formation and create de novo R-loops in constructs that do not show initiation without nicks.
Cas9 target site identification in VR inserts #
Primary requirement for specific Cas9 activity is the presence of a PAM site downstream of the target site. To identify these sites in VR insert constructs I created a snakemake workflow: VR-Cas9. The workflow uses the complete VR insert constructs (currently just the T7 initiation series) and determines suitable target sites and generates oligos.
VR-Cas9 workflow designs guide RNAs for use with NEB sgRNA synthesis kit
As of 12/9/21 the workflow designs oligos that are compatible with NEB NEB sgRNA synthesis kit. These oligos should not be used for other sgRNA synthesis approaches.
In addition to describing the target sequences and generate NEB kit compatible oligos the workflow will also generate maps of each plasmid showing the selected target sites.
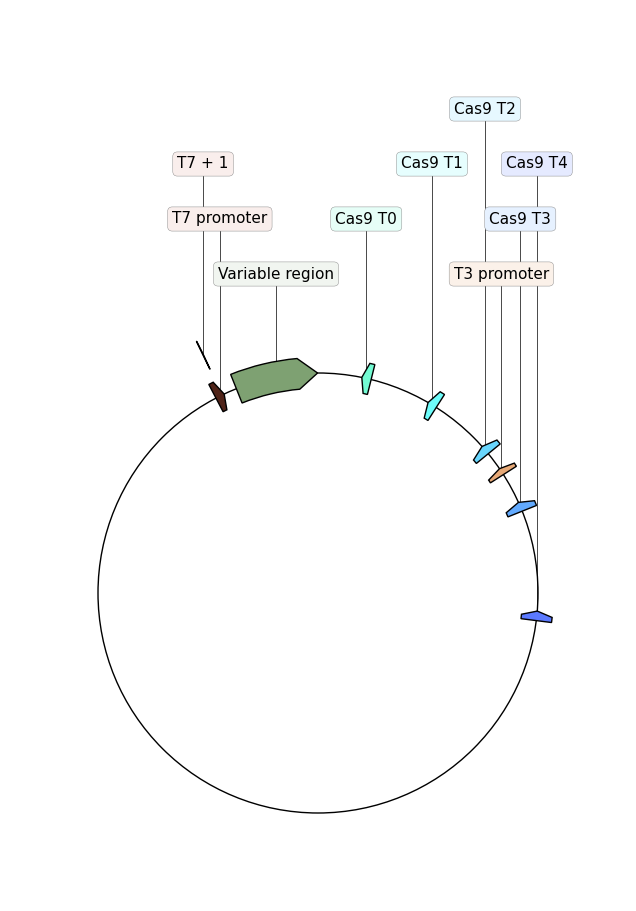
Updated versions of the VR-Cas9 workflow (as of 2/1/22) do not produce this
type of plasmid map.2/1/22 #
Update VR-Cas9 workflow: bug fixes and smarter target finding #
Over the last two days I made updates to the workflow that both fixed
bugs and will hopefully select better targets. The main bug fixed / feature
added was re-slicing plasmids to linear sequence based on specific feature
locations. Previously the workflow just used the linear script as provided
by the genbank file. This is fine if you want to distribute sgRNAs all over
the plasmid but not if you want to avoid specific regions or if the distance
of the target sequence to the start is important (as is this case). I also
implemented Doench et al 2014 on target efficiency score as the primary
target selector. The plot below shows on-target scores of recently designed
sgRNAs that were selected by the pipeline vs. not selected vs. the ones I
originally designed (labeled as excluded in this plot).
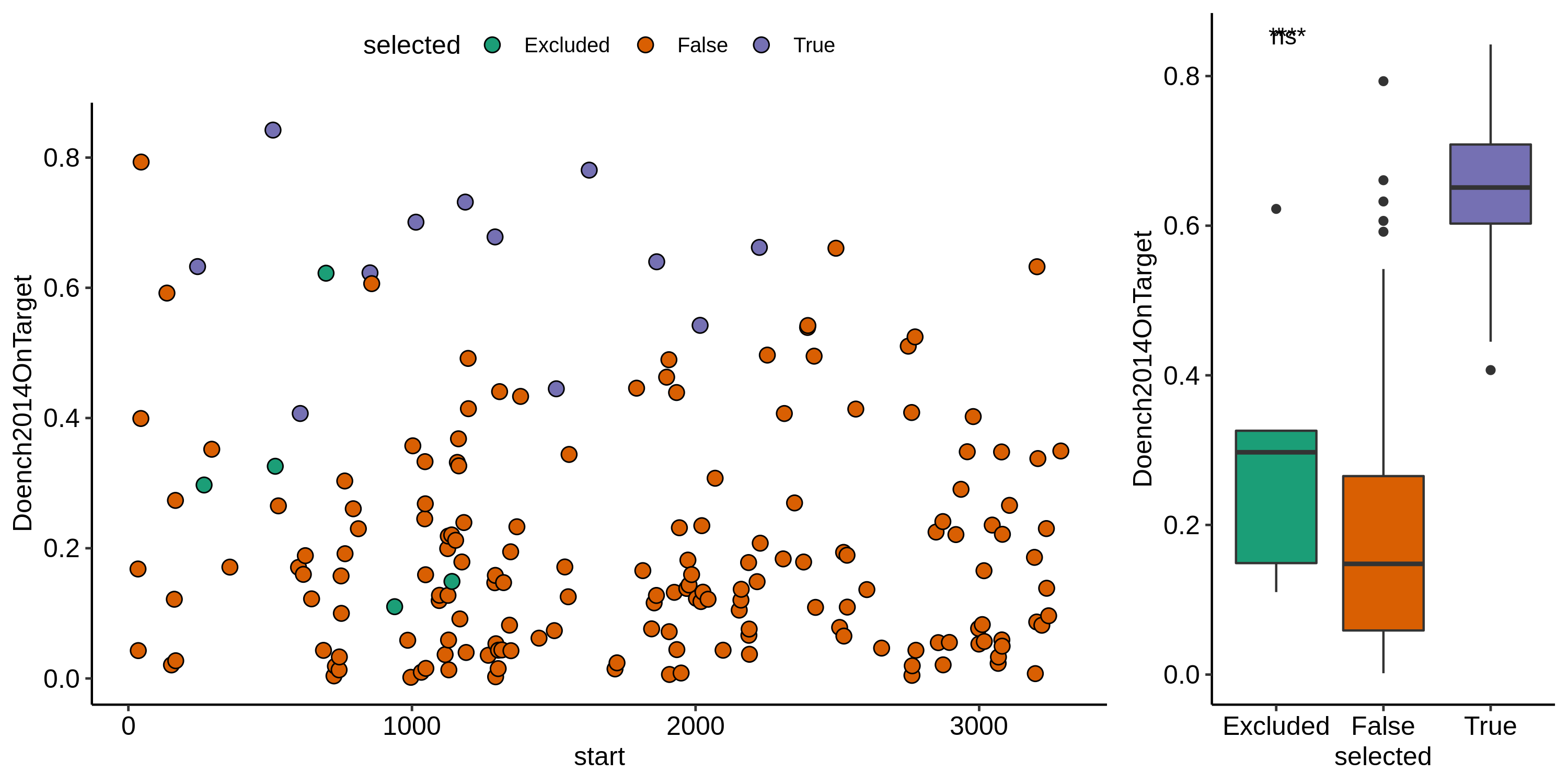
sgRNAs produced by the updated pipeline in general have much better on-target scores which will hopefully mean less sgRNA optimization is required. Additionally I removed target sequences that overlaped with the variable region so any sgRNAs from future runs using this same masking should be universal for all templates.I went ahead and ordered the first 5 oligos (from the start of sequence, oligo ID oEH25-29) from IDT. Afterwards (probably should have done before) I compared the nicking efficiencies of my current sgRNAs (oligo IDs oEH15-19) to the predicted on-target score, shown in the plot below.
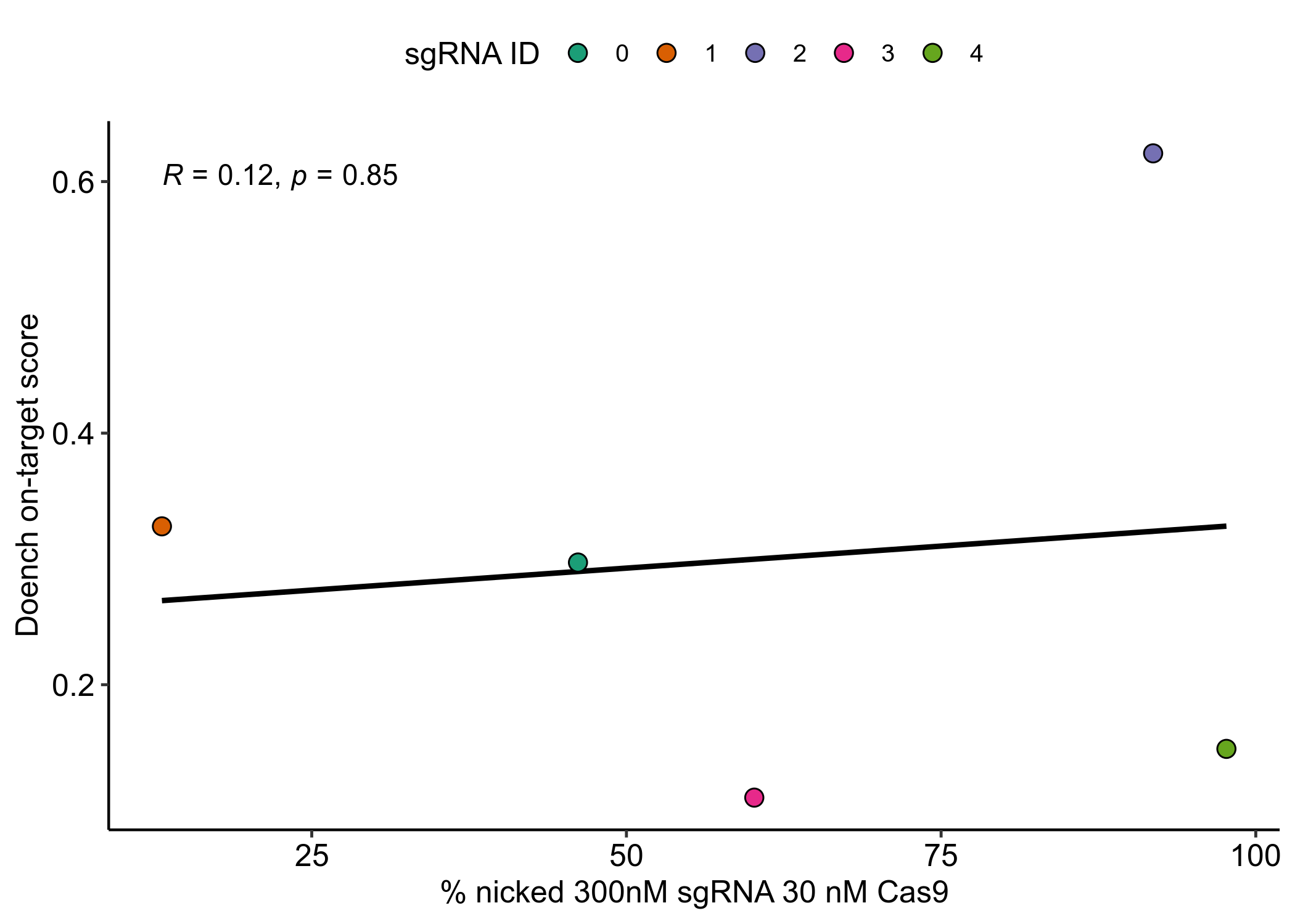
On-target efficiency corresponds poorly to actual sgRNA performance, at least
for these five sgRNAs. This may be for a number of reasons including that
most Cas9 metrics are designed for in-vivo approaches and the face this
is a very small sample size. For now some metric is better than no metric. I
may update this method in the future if the sgRNAs designed with optimizing
Doench et al 2014 on-target scores do not show general improvement over
the first batch.
Guide RNA synthesis #
Once guide RNAs are selected for a specific construct(s) I will need to get them into a tube with Cas9 nickase somehow.
A note on guide RNAs
There is a difference between gRNA and sgRNA when used in the context of Cas9. gRNA refers to the target specific sequence while sgRNA single guide RNA) refers to the gRNA and the scaffold RNA that interacts with the Cas9 protein structure. More details from the addgene guide.
Ordering sgRNAs from IDT is expensive (around $200 per guide) which is not ideal. Alternative would be to order DNA sequence of the target RNA and then clone into a sgRNA expression vector. I would then PCR amplify the sgRNA region, transcribe in vitro, and then extract RNA to use with the Cas9 nickase.
Would need to get expression vector; pT7-gRNA example shown in the image below.
Alternatively, Fred suggested ordering the NEB sgRNA synthesis kit which compresses everything into one tube; would just need to order DNA oligos then which I think will be the way to go.
1/18/22 #
Guide RNA oligo and synthesis kit order #
Ordered 5 of the 16 oligos designed by VR-Cas9 workflow for pFC9VR13 template, along with the NEB sgRNA synthesis kit and EnGen Spy Nickase. Oligo sequences and details are available in the Ethan’s oligos sheet at this link.