Background: Why a round two is needed #
While previous midi preps of this series were successful they were slowly nicked overtime due to resuspension in 10 mM TrisHCL. I came up with a plan to repair these nicks to avoid having to redo all 31 midi preps by intentially nicking samples with BspQI nicking endonuclease, ligating nicks and then re-supercoiling with DNA gyrase but evaluation of this approach showed it would not be feasible without significant optimization. So I am forced to repeat all preps and pray that the plasmid recombination issues that plagued the first round do not rear their head(s) again.
Re-transforming all T7Init plasmids #
2/15/22 #
Since previous transformations are about 2 months old at this point I started by re-transforming previously sequenced midi prep samples using my homemade chemically competent cells. I followed NEB transformation protocols for timings and temperatures. See table below for complete description of transformations.
I incubated plates in 37 room overnight for pickup the next morning.
2/16/22 #
Picked up plates at 8:40 AM the next day (~20 hours incubation time) and found all plates had colonies (between 75 and 200+) per plate except VR1 plate which had around 25 colonies. Negative control (no plasmid in transformation) had no colonies. Placed all plates in the deli fridge for long term storage.
Batch 1: Midi preps T7InitVR1-16 #
2/16/22 #
Prepared 4L of 1x LB, autoclaved, cooled and then added 100 ug / ml AMP. Dispensed ~200 ml of LB under flame into 17 autoclaved 1 L flasks. I then innoculated each flask with 1 colony from T7Init transformation plates (VR1-16) using pipette tips and used the last flask as a negative control (pippete tip but no colony). I placed all flasks into various second floor shakers and incubated at 37C with shaking at ~200 rpm starting at 5:40 PM.
2/17/22 #
Prep execution #
Picked up flasks at 6:43 AM and found all cultures had grown and negative control showed no signs of growth. Proceeded with lab midi prep protocol on all samples.
A note on spinning down protein lysate in 500 ml bottles
It seems for whatever reason that protein lysate does not separate as well from supernatent when spinning down in 500 ml bottles (surface area?). I had to spin samples in 500 ml bottles for twice as long as 250 ml bottles (20 mins) at 12000 rpm (2000 rpm faster than 250 ml) to get decent separation.
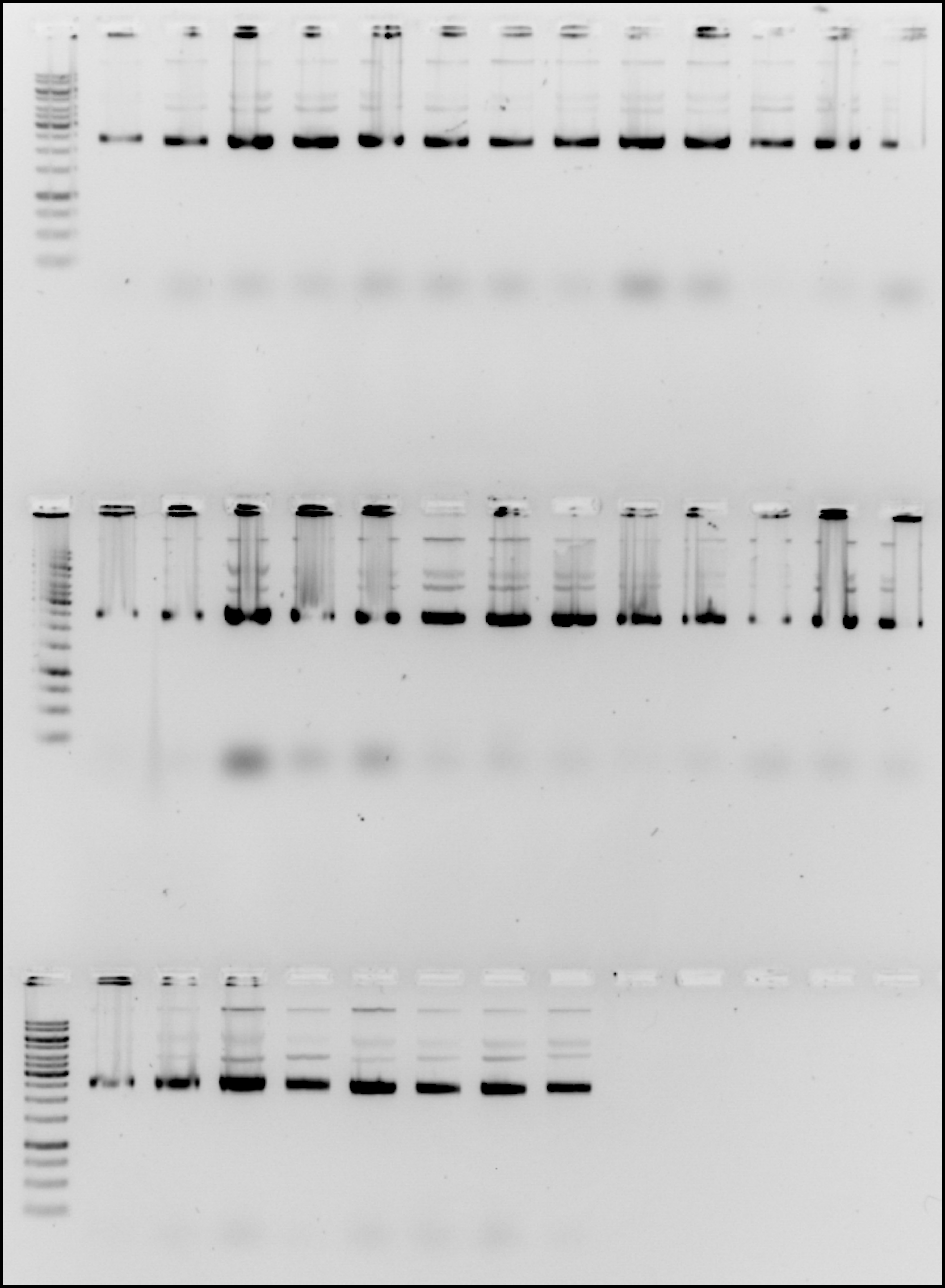
After completing LiCl cleanup ran 0.5 ul of samples on an agarose gel which is shown in the image below.
Overall preps looked pretty good. No evidence of plasmid recombination. Sample VR14 looks either relaxed or nicked and VR1 looks like I lost the pellet or something but 14/16 is not bad.
Batch 2: Midi prep T7InitVR16-31 preparation #
While working on midi preps also prepared another 4L of culture and washed and autoclaved 1 L flasks. After completing preps dispensed 200 ml of LB + AMP into flasks under flame and inoculated with single colonies from VR17-31 plates plus a colony from VR11. Incubated cultures at 37C with shaking at 200 rpm in various second floor shakers.
Somehow I got it in my head I needed to redo VR11 midi prep. Not sure what I was thinking. That is why next preps include a VR11 sample instead of 1 or 14.
Batch 3: Midi preps T7InitVR17-31 + VR11 #
2/18/22 #
Prep execution #
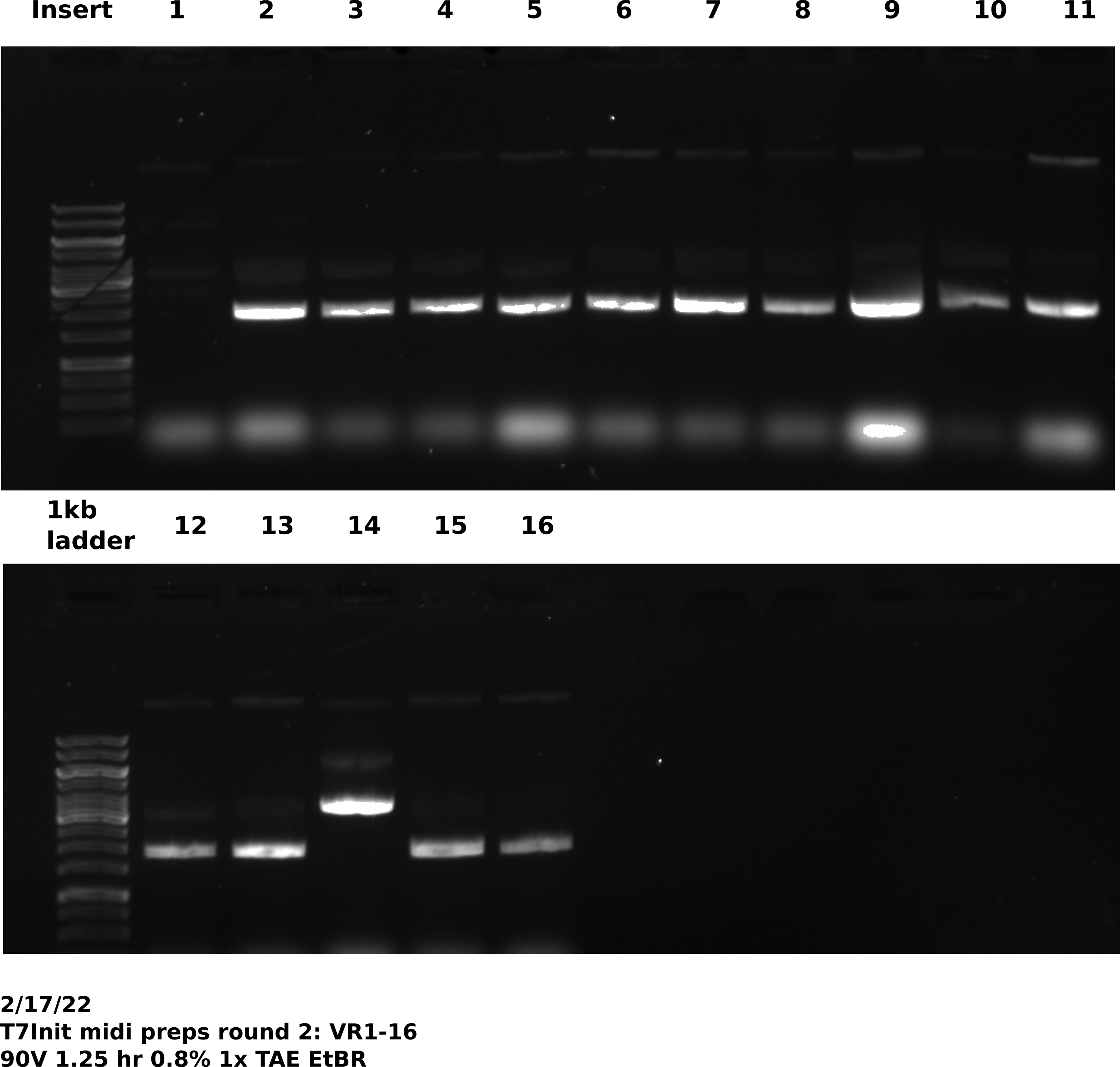
Picked up cultures at 7:20 AM and found all cultures had grown and negative control showed no signs of growth. Proceeded with lab midi prep protocol on all samples. After completing LiCl purification of all samples ran 0.5 ul of each sample out on a gel, shown in the image below.
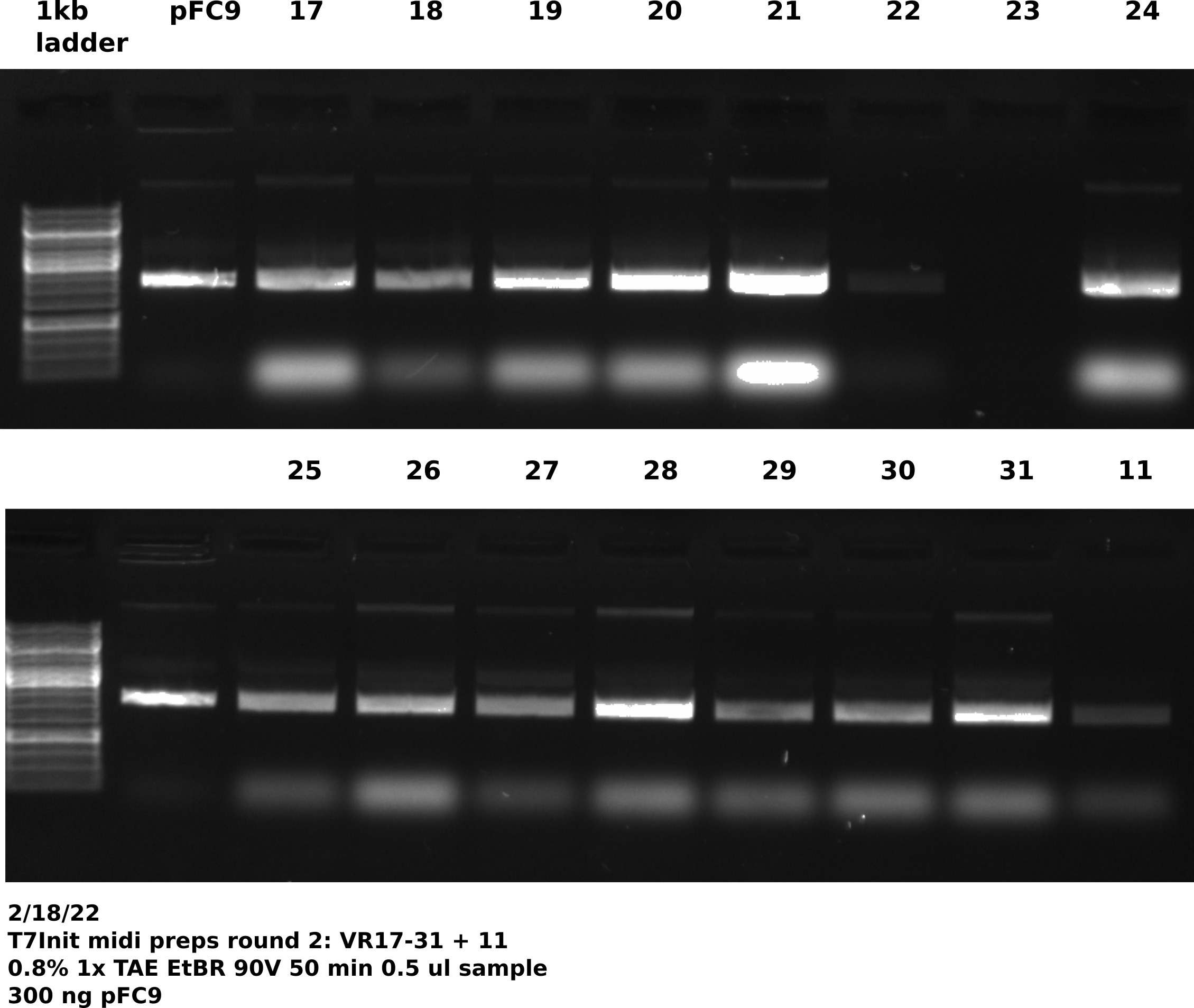
Tada’s loaded this gel with multi-channel pipette. VR23 appeared to have no DNA at all. So I had him spec the sample and it showed too concentrated to measure. I think there is DNA in there but he just missed the well.
Overall things again seem to be looking good and as expected.
Sample prep for Sanger sequencing #
Prepared all current midi preps for sanger sequencing VR2-31 using 4 ul of each midi prep. Complete description of reactions and samples in the table below.
Analysis of Sanger seq data #

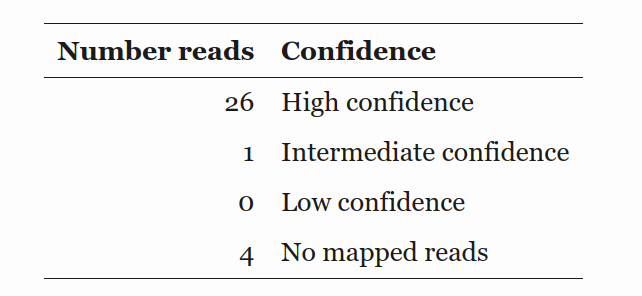
VR31 read is short due to high G region #
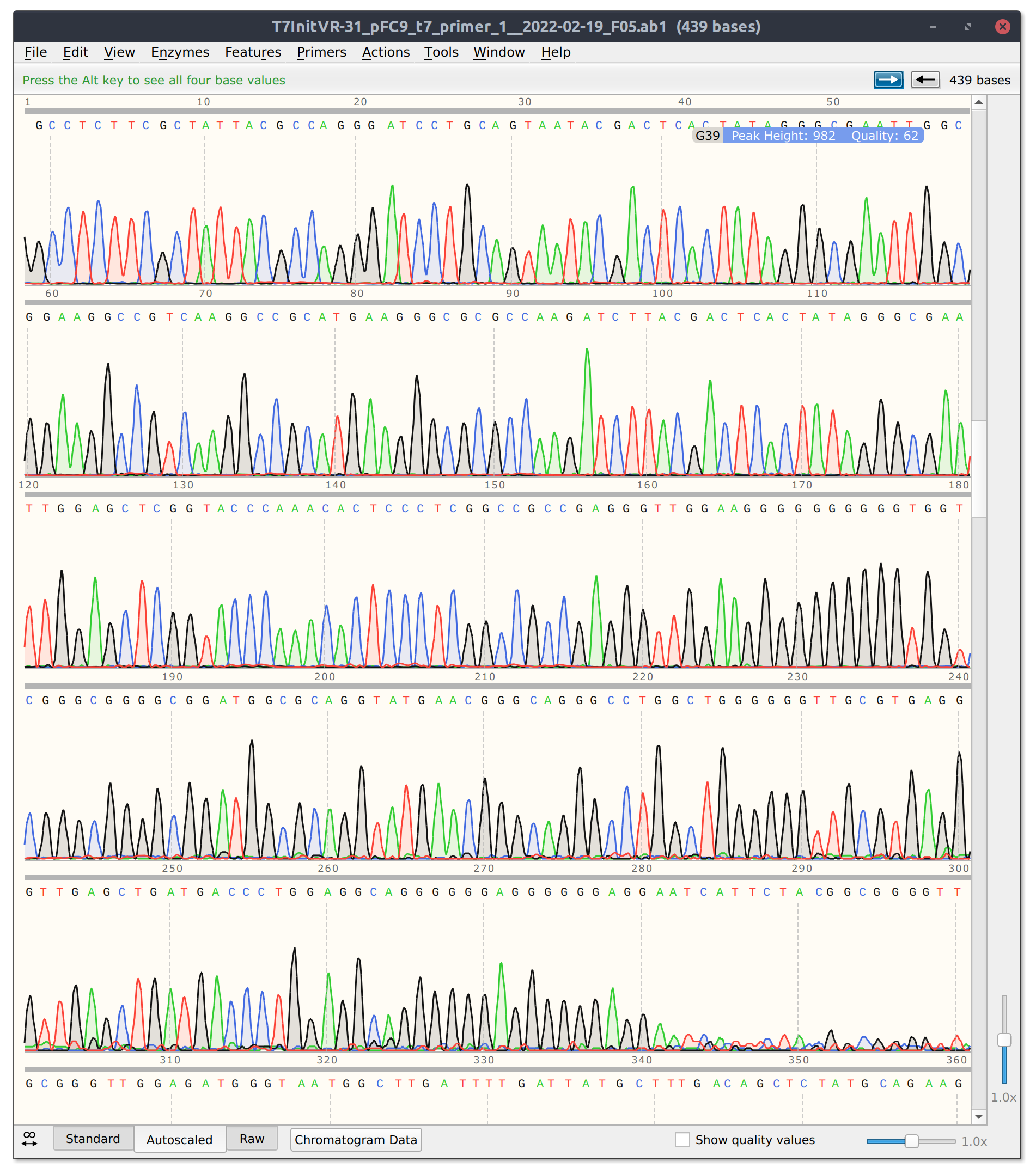
Batch 4: Midi preps VR 1, 10, 12, 17, and 22 #
2/22/22 #
Trouble-maker insert re-transformations #
Aiming to finish off midi preps with these last few. Since these guys have been such buggers I re-transformed all of these insert containing plasmids and plated 300 ul of the transformation mix and incubated overnight using homemade chemically competent cells.
RnaseA cleanup assessment #
RNA contamination is a problem with basically all midi preps. I used pFC9VR16 to test what kind of RnaseA digestion would be required to remove most of the RNA in midi samples.
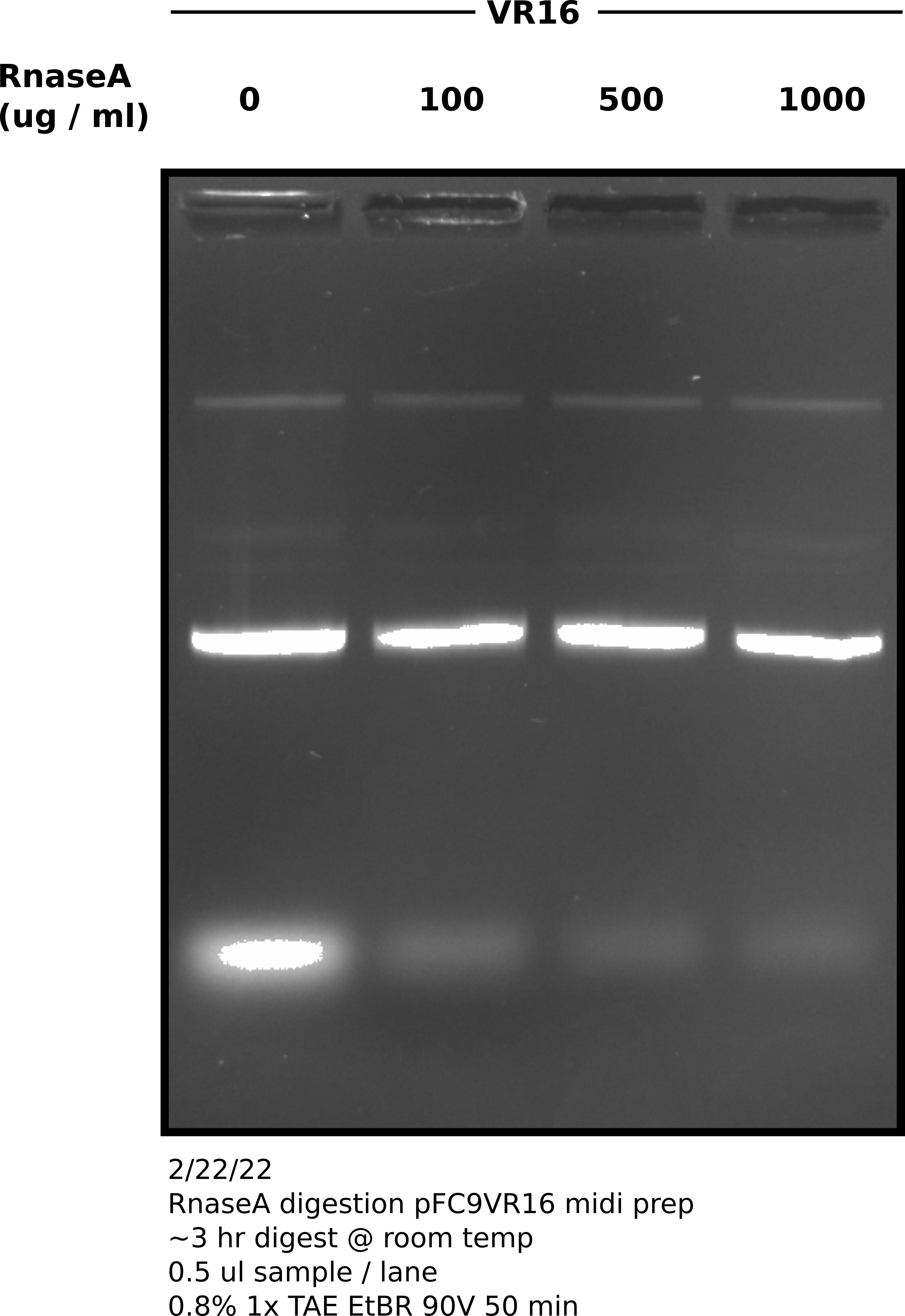
1000 ug / ml was the most successful over the course of the three hour digestion. Moving forward with this concentration but I think increasing the digestion time would be beneficial.
2/23/22 #
Midi prep prep #
Picked up plates and all had a significant number of colonies (probably only needed to plate 100 ul or so of transformation mix) and the negative control had 1 colony (clean enough).
I placed all plates in the fridge and prepared 2 L of 1x LB + 100 ug/ml AMP. Dispensed LB into autoclaved 1L flasks and inoculated with 1 colony from plates transformed yesterday. Placed flasks on various second floor shakers and incubated at 37C with shaking at 200 rpm to pick up in the morning.
Innoculated a total of 6 samples, VR10, 17, 2 colonies from VR1 and colonies from VR22C4 and VR22C4.
RnaseA digestion of existing preps #
In order to remove RNA contamination from existing Sanger sequencing verified preps I added 50 ug of 10 mg/ml RnaseA (final concentration 1kug / ml) and incubated at room temperature starting at 11 AM. Digesting samples 2-9, 11-16, 19-21, 23-31. Following RnaseA digestion will precipitate with Phenol-Chloroform EtOH and re-suspend in TE buffer.
Placed samples back into freezer at 5:00 PM, did not do any precipitation. Waiting to run aliquots of a few of the samples on gel to access if further digestion is required.
2/24/22 #
Midi prep execution #
Picked up cultures started yesterday and while some grew well overnight to saturation others appear to still be in the log phase of growth. OD was higher than control and cells where growing but not at saturation. Samples 17 and 10 reached saturation, 1B and 22C7 where almost there and 22C4, 1A and 12 needed to grow for longer. Proceeded with prep of VR10, 17, 1B and 22C7 and allowed 1A and 22C4 to continue growing at 37C shaking at 200 rpm.
Normal number of cells present for VR10 and 17 cultures, about 1/4th that for 22C7 and 1B. Not enough DNA to continue with the later two after isopropyl precipitation. VR10 and 17 were successful.
Comparison of culture growth density #

After completing VR10 and 17 preps checked on VR22C4, 1A and 12 and all had grown to saturation. Plates from these colonies where much smaller so might have just needed more time to grow. Completed isopropyl precipitation with Aiden around 5 PM, re-suspended pellet in 0.5 TE skipping over RnaseA digestion and added 0.5 ml LiCl. Placed in freezer to incubate overnight.
2/25/22 #
Sanger seq sample submission #
Submitted the following samples to Quintara for sequencing.
| Backbone | Insert Number | Primer | Sample Name | Prep date |
|---|---|---|---|---|
| pFC9 | VR12 | pFC9_t7_primer_1 | pFC9VR12 | 12/24/22 |
| pFC9 | VR22C4 | pFC9_t7_primer_1 | pFC9VR22C4 | 12/24/22 |
| pFC9 | VR1 | pFC9_t7_primer_1 | pFC9VR1 | 12/24/22 |
| pFC9 | VR31 | VRI_primer_2 | pFC9VR31 | 12/18/22 |
I forgot to include samples I prepped earlier in the day yesterday, so these are just the ones I completed with Aiden plus VR31 sequenced in the reverse direction to bypass very G rich sequence prone to killing reads.
Analysis of 2/25 sequencing samples #
Downloaded reads from Quintara and added to VR-verify workflow. All reads aligned with high confidence except VR-22 which had intermediate confidence due to short read length. However, upon looking at traces I noticed all of the reads that used pFC9_t7_primer_1 had short read lengths in comparison to other sample submissions with signal dying about 400 bp out. This primer is running very low, may be scraping the bottom of the barrel and need to re-order.
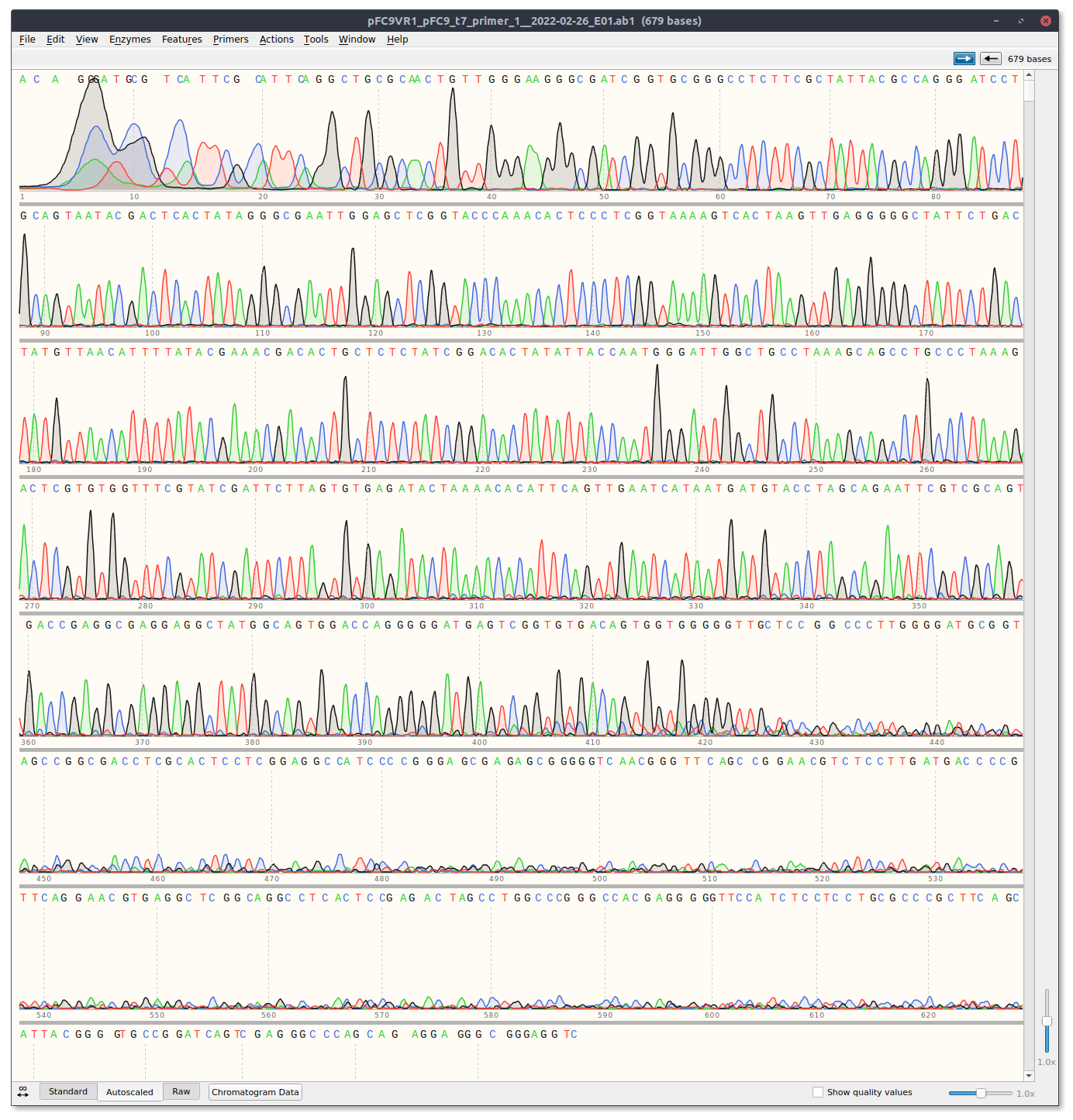
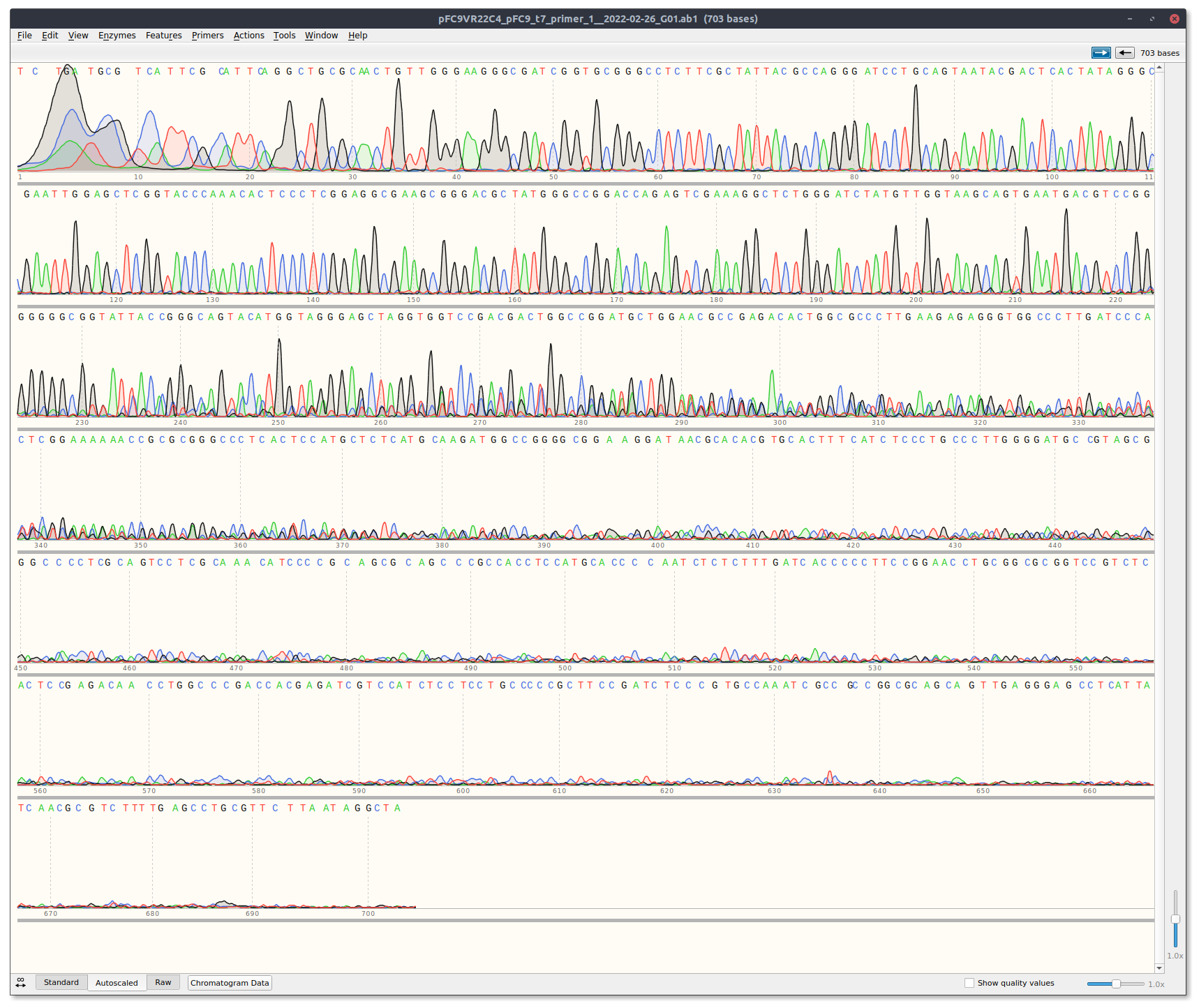
VR1 and VR22C4 are both short, with VR22C4 being very short.
VR1 Trace #
VR22 Trace #
VR31 Trace (Did not use pFC9_t7_primer_1) #
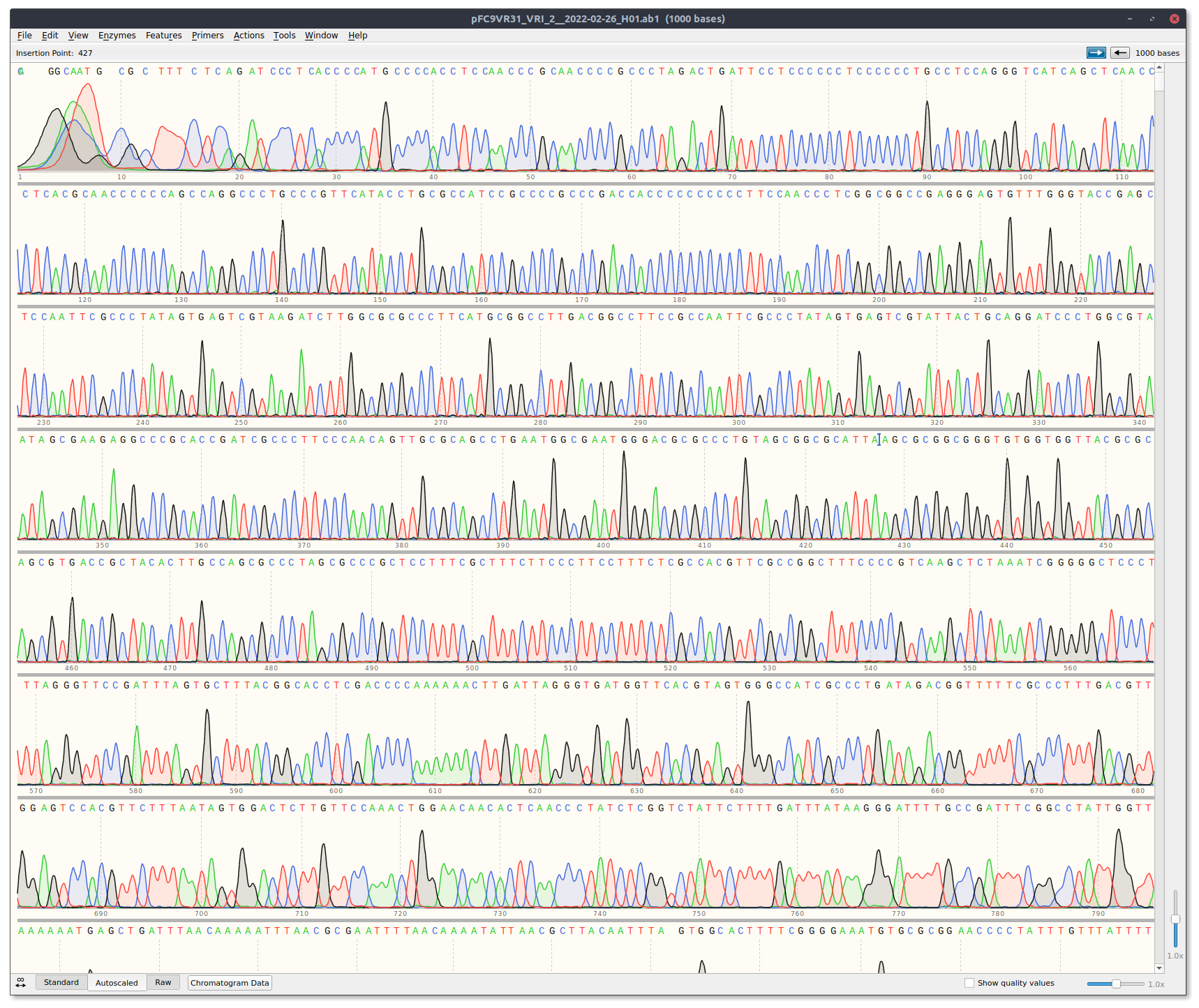
QuBit sample concentration measurements #
While RnaseA digestion has not removed 100% of the RNA in midi samples and this interfers with OD measurements, QuBit uses an intercalating dye which should only measure dsDNA concentrations. Because of this I am using QuBit to determine sample concentrations of existing RnaseA digested midi-preps.
2/25/22 #
Measuring existing midi prep samples. All measured samples where digested with RnaseA for at least 4 hours at room temperature and then phenol-chloroform precipitated using homemade phase lock columns.
Sample measurements #
Complete table available at this link.
Sample measurement table
| Sample Name | Sample Date | Sample volume measured (ul) | Dye Volume (ul) | Concentration (ng/ul) | Measurement Date | Measurement mode |
|---|---|---|---|---|---|---|
| pFC9VR2 | 2/17/22 | 1 | 199 | 826 | 2/25/22 | dsDNA Broad Range |
| pFC9VR3 | 2/17/22 | 1 | 199 | 772 | 2/25/22 | dsDNA Broad Range |
| pFC9VR4 | 2/17/22 | 1 | 199 | 688 | 2/25/22 | dsDNA Broad Range |
| pFC9VR5 | 2/17/22 | 1 | 199 | 734 | 2/25/22 | dsDNA Broad Range |
| pFC9VR6 | 2/17/22 | 1 | 199 | 672 | 2/25/22 | dsDNA Broad Range |
| pFC9VR7 | 2/17/22 | 1 | 199 | 708 | 2/25/22 | dsDNA Broad Range |
| pFC9VR8 | 2/17/22 | 1 | 199 | 576 | 2/25/22 | dsDNA Broad Range |
| pFC9VR9 | 2/17/22 | 1 | 199 | 650 | 2/25/22 | dsDNA Broad Range |
| pFC9VR10 | 2/24/22 | 1 | 199 | 882 | 2/25/22 | dsDNA Broad Range |
| pFC9VR11 | 2/18/22 | 1 | 199 | 836 | 2/25/22 | dsDNA Broad Range |
| pFC9VR13 | 2/17/22 | 1 | 199 | 644 | 2/25/22 | dsDNA Broad Range |
| pFC9VR14 | 2/17/22 | 1 | 199 | 774 | 2/25/22 | dsDNA Broad Range |
| pFC9VR15 | 2/17/22 | 1 | 199 | 660 | 2/25/22 | dsDNA Broad Range |
| pFC10VR16 | 2/17/22 | 1 | 199 | 808 | 2/25/22 | dsDNA Broad Range |
| pFC9VR17 | 2/24/22 | 1 | 199 | 754 | 2/25/22 | dsDNA Broad Range |
| pFC9VR18 | 2/18/22 | 1 | 199 | 960 | 2/25/22 | dsDNA Broad Range |
| pFC9VR19 | 2/18/22 | 1 | 199 | 852 | 2/25/22 | dsDNA Broad Range |
| pFC9VR20 | 2/18/22 | 1 | 199 | 896 | 2/25/22 | dsDNA Broad Range |
| pFC9VR21 | 2/18/22 | 1 | 199 | 758 | 2/25/22 | dsDNA Broad Range |
| pFC9VR23 | 2/18/22 | 1 | 199 | 682 | 2/25/22 | dsDNA Broad Range |
| pFC9VR24 | 2/18/22 | 1 | 199 | 754 | 2/25/22 | dsDNA Broad Range |
| pFC9VR25 | 2/18/22 | 1 | 199 | 820 | 2/25/22 | dsDNA Broad Range |
| pFC9VR26 | 2/18/22 | 1 | 199 | 668 | 2/25/22 | dsDNA Broad Range |
| pFC9VR27 | 2/18/22 | 1 | 199 | 792 | 2/25/22 | dsDNA Broad Range |
| pFC9VR28 | 2/18/22 | 1 | 199 | 674 | 2/25/22 | dsDNA Broad Range |
| pFC9VR29 | 2/18/22 | 1 | 199 | 592 | 2/25/22 | dsDNA Broad Range |
| pFC9VR30 | 2/18/22 | 1 | 199 | 656 | 2/25/22 | dsDNA Broad Range |
| pFC9VR31 | 2/18/22 | 1 | 199 | 1020 | 2/25/22 | dsDNA Broad Range |
Accurate DNA measurements using the QuBit with high concentration samples
The measurements shown in the chart above all used 1 ul of midi prep samples and the
broad rangesetting. After attempting to run a gel with all midi preps with an equal mass of each sample per lane based on the measurements above I found sample concentrations were not accurate as some lanes had significantly more DNA than others.It turns out that you need a different dye to use the broad range setting. To get around this highly concentrated samples should be diluted first, then measured using the
1x dsDNA high sensitivitysetting.
3/4/26 #
Given what I learned about QuBit usage and the requirement for specific dyes
for specific settings I re-measured sample concentrations for verified and
RnaseA digested T7Init midi prep samples by first diluting 1 ul of each sample
in 49 ul TE and then using 1 ul of this dilution with 199 ul
1x dsDNA high sensitivity QuBit dye.
Sample measurements and stock concentration calculations are shown in the table below and are also available at this link.
Updated QuBit sample concentrations
| Construct | Sample Date | measurement | Stock concentration |
|---|---|---|---|
| pFC9VR-1 | 2/24/22 | 11.7 | 585 |
| pFC9VR-2 | 2/17/22 | 8.72 | 436 |
| pFC9VR-3 | 2/17/22 | 9.04 | 452 |
| pFC9VR-4 | 2/17/22 | 3.46 | 173 |
| pFC9VR-5 | 2/17/22 | 5.8 | 290 |
| pFC9VR-6 | 2/17/22 | 5.18 | 259 |
| pFC9VR-7 | 2/17/22 | 9.44 | 472 |
| pFC9VR-8 | 2/17/22 | 2.28 | 114 |
| pFC9VR-9 | 2/17/22 | 4.46 | 223 |
| pFC9VR-10 | 2/24/22 | 15.3 | 765 |
| pFC9VR-11 | 2/18/22 | 8.98 | 449 |
| pFC9VR-12 | 2/24/22 | 11 | 550 |
| pFC9VR-13 | 2/17/22 | 14.2 | 710 |
| pFC9VR-14-1 | 3/6/22 | 4.22 | 211 |
| pFC9VR-15 | 2/17/22 | 3.88 | 194 |
| pFC9VR-16 | 2/17/22 | 4.78 | 239 |
| pFC9VR-17 | 2/24/22 | 8.82 | 441 |
| pFC9VR-18 | 2/18/22 | 7.14 | 357 |
| pFC9VR-19 | 2/18/22 | 7.7 | 385 |
| pFC9VR-20 | 2/18/22 | 16.9 | 845 |
| pFC9VR-21 | 2/18/22 | 23.8 | 1190 |
| pFC9VR-22 | 2/24/22 | 18.2 | 910 |
| pFC9VR-23 | 2/18/22 | 7.08 | 354 |
| pFC9VR-24 | 2/18/22 | 8.84 | 442 |
| pFC9VR-25 | 2/18/22 | 16.6 | 830 |
| pFC9VR-26 | 2/18/22 | 6.6 | 330 |
| pFC9VR-27 | 2/18/22 | 14.5 | 725 |
| pFC9VR-28 | 2/18/22 | 12.7 | 635 |
| pFC9VR-29 | 2/18/22 | 10.8 | 540 |
| pFC9VR-30 | 2/18/22 | 11.1 | 555 |
| pFC9VR-31 | 2/18/22 | 21.1 | 1055 |
Overall concentrations are quite a bit lower than previous results; but with 0.5 ml of each sample there is more than enough for all required experiments but maybe not enough for eternity.
Final round up #
3/6/22 #
Re-ran VR-verify pipeline with all current Sanger reads of T7Init round 2 midi preps.
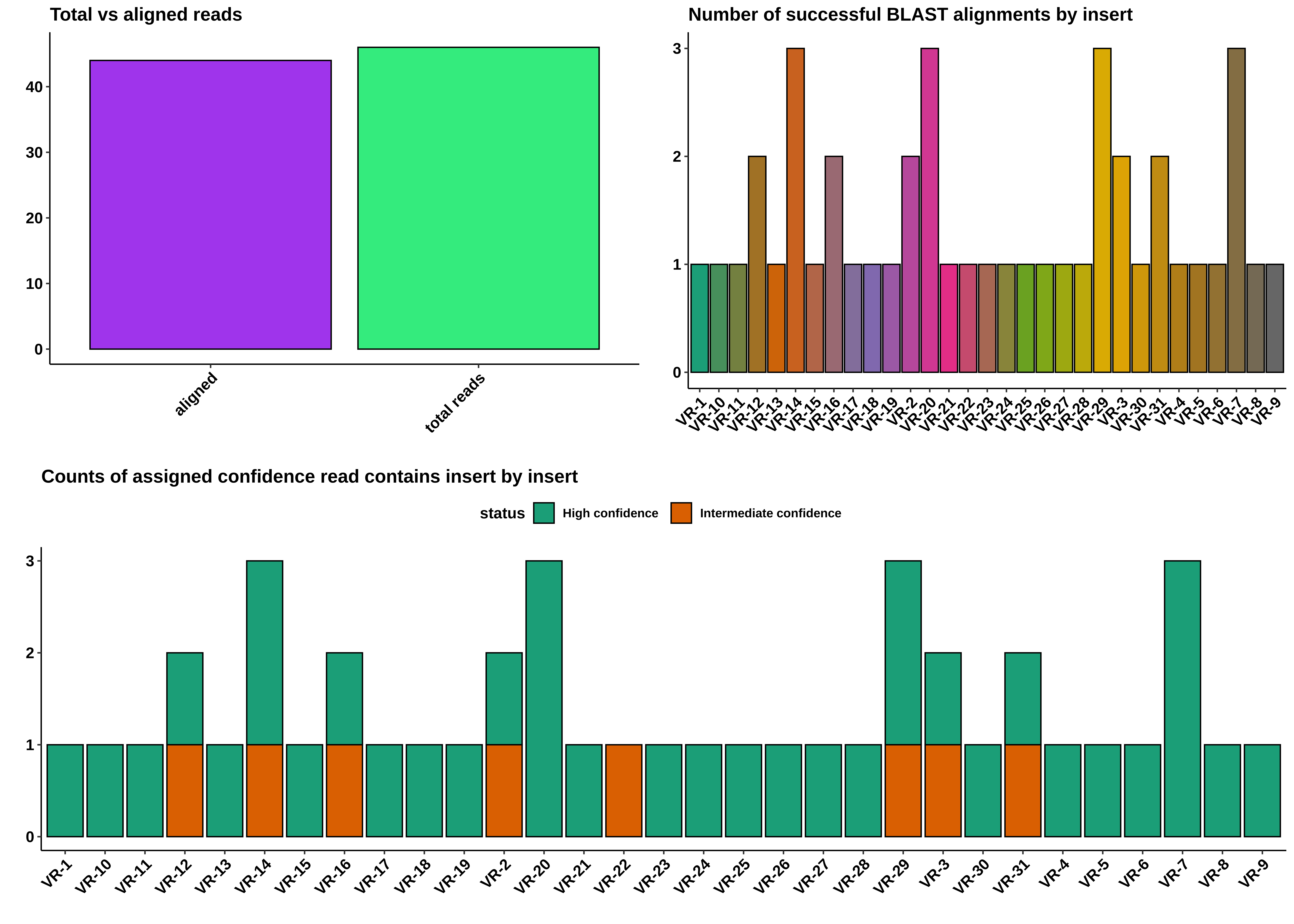
All inserts have at least 1 high confidence read except for VR-22 due to a short read. This construct looks normal on a gel so I am counting it because aligned to the insert region is good.
Pipeline output, configuration and sequencing data can be downloaded from this link.
Family picture #
3/2/22 #
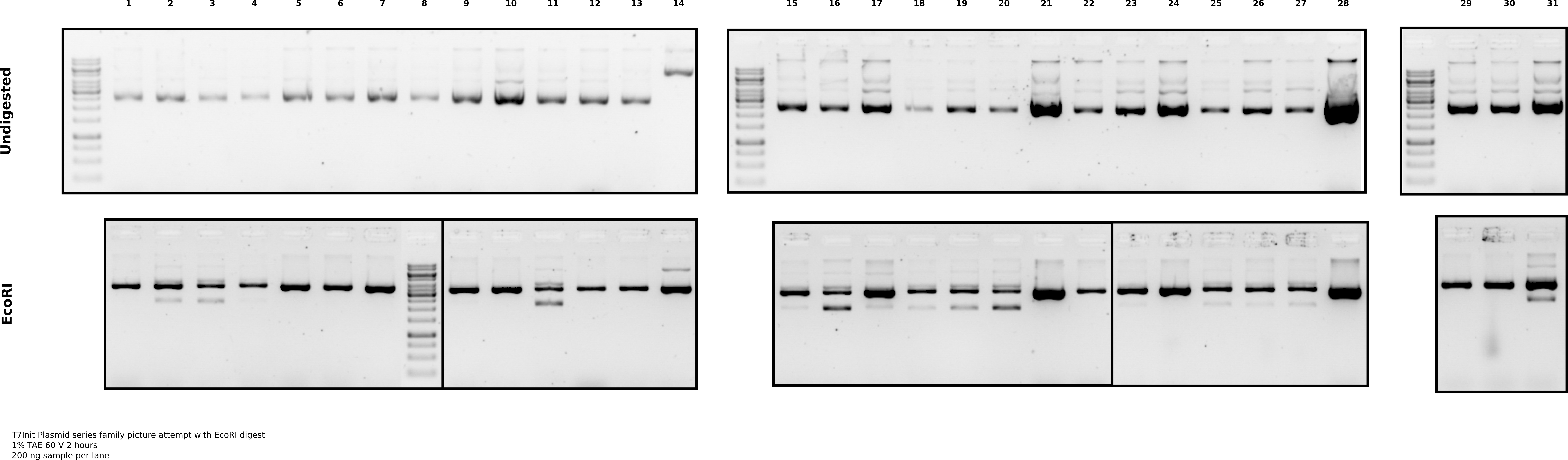
3/7/22 #
Gel layout
| Construct | Construct mass (ng) | Lane |
|---|---|---|
| 1 kb ladder (O’gene ruler) | 1 ul | 1 |
| pFC9 | 100 | 2 |
| pFC9VR1 | 100 | 3 |
| pFC9VR2 | 100 | 4 |
| pFC9VR3 | 100 | 5 |
| pFC9VR4 | 100 | 6 |
| pFC9VR5 | 100 | 7 |
| pFC9VR6 | 100 | 8 |
| pFC9VR7 | 100 | 9 |
| pFC9VR8 | 100 | 10 |
| pFC9VR9 | 100 | 11 |
| pFC9VR10 | 100 | 12 |
| pFC9VR11 | 100 | 13 |
| pFC9VR12 | 100 | 14 |
| 1 kb ladder (O’gene ruler) | 1 ul | 16 |
| pFC9 | 100 | 17 |
| pFC9VR13 | 100 | 18 |
| pFC9VR14 | 100 | 19 |
| pFC9VR15 | 100 | 20 |
| pFC9VR16 | 100 | 21 |
| pFC9VR17 | 100 | 22 |
| pFC9VR18 | 100 | 23 |
| pFC9VR19 | 100 | 24 |
| pFC9VR20 | 100 | 25 |
| pFC9VR21 | 100 | 26 |
| pFC9VR22 | 100 | 27 |
| pFC9VR23 | 100 | 28 |
| 1 kb ladder (O’gene ruler) | 1 ul | 30 |
| pFC9 | 100 | 31 |
| pFC9VR24 | 100 | 32 |
| pFC9VR25 | 100 | 33 |
| pFC9VR26 | 100 | 34 |
| pFC9VR27 | 100 | 35 |
| pFC9VR28 | 100 | 36 |
| pFC9VR29 | 100 | 37 |
| pFC9VR30 | 100 | 38 |
| pFC9VR31 | 100 | 39 |
Gel parameters
| Parameter | Value |
|---|---|
| Agarose % | 1 |
| Buffer | 1x TAE |
| EtBr | 1 ul / 100 ml |
| Voltage | 60 |
| Runtime (min) | 120 |
| Run date | 3/7/22 |
Gel image #