TacT1T2 initiation series plasmids #
This series of plasmids utilizes the TacTIT2 backbone and the T7 initiation series to insert initiation series variable regions upstream of the T1T2 sites to
- Take advantage of predictable termination for single round transcription assays.
- Utilize the Tac (E. coli promoter) to use a more complex polymerase compared to T7 or T3.
Tada’s is taking the lead on creating these constructs utilizing pFC8TacT1T2 plasmid as the backbone. pFC8TacT1T2’s identity was confirmed using Sanger sequencing.
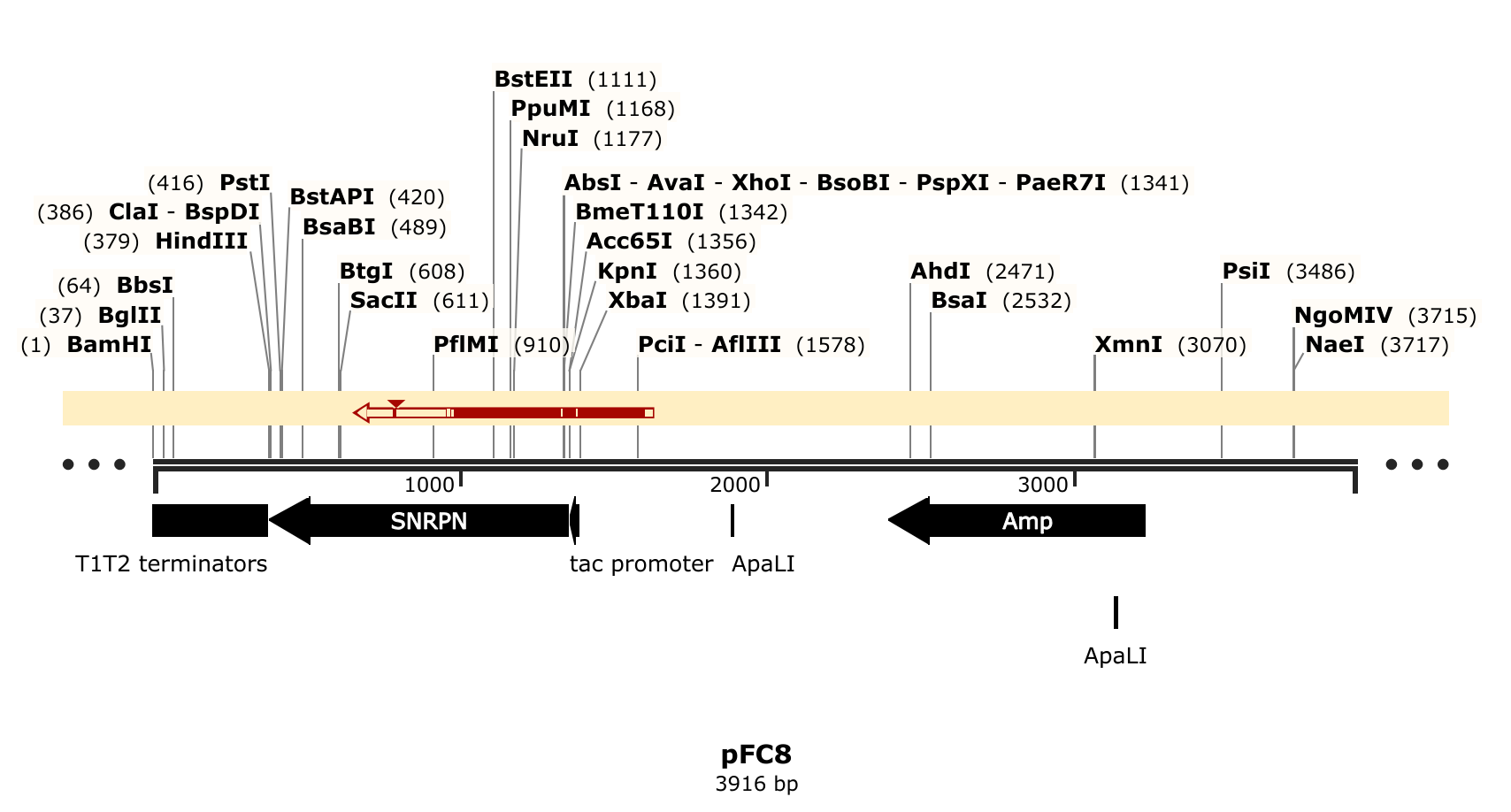
Sequence and trace files available at this link.
However, the gene of interest carried in the existing backbone does not actually matter to the creation of the final construct as it is completely removed via a HindIII, KpnI double digest.
Identification by Sanger sequencing #
Sequence potential constructs using pFC8tac_tac_promoter_Primer_2. This primer sequences reverse strand and binds downstream of the insert location.
Review of Tada’s 2/11/22 mini preps #
2/13/22 #
Tada’s submitted a number of samples for sequencing on Friday and he downloaded reads from quintara on Saturday. I downloaded on Sunday to run through the VR-verify pipeline but when I did I found that none of the reads aligned. Upon comparing the files we downloaded to each other (both from the same link on Quintara page) the reads were completely different.
I BLASTed the reads I downloaded and then analyized the results in this jupyter notebook. While some reads did not align to anything at all, many others aligned to yeast genome and various expression vectors. The alignment results are shown in the table below.
There is no way the primers Tada’s actually used for sequencing could get reads from these templates. This is telling me there was a data management issue on Quintara’s end. Will call later to follow up.