pFC53tac and pFC8tac digests plus primer design #
Today I am continuing to work on determining the true identity and content of the pFC53 plasmids. I also designed primers to amplify each plasmid for sequencing.
pFC53tac and pFC8tac digest #
I digested Rachel’s pFC53tac and pFC8tac and my own pFC53tac I prepared from Rachel’s pFC53tac stock. Below are the expected results for digests of true pFC53tac and pFC8tac.

MW: 1 kb DNA Ladder
1: pFC53tacT1T2
1. 4320 bp
2: pFC53tacT1T2
BamHI + NotI
1. 4299 bp
2. 21 bp
3: pFC53tacT1T2
EcoRI
1. 4320 bp
4: pFC8tacT1T2
1. 3916 bp
5: pFC8tacT1T2
BamHI + NotI
1. 3306 bp
2. 589 bp
3. 21 bp
6: pFC8tacT1T2
EcoRI + XbaI
1. 2574 bp
2. 818 bp
3. 524 bp
Results #
Ran 0.08 agarose gel in TAE for 45 mins at 120V (shown below). R are Rachel’s plasmids while EH are my own.
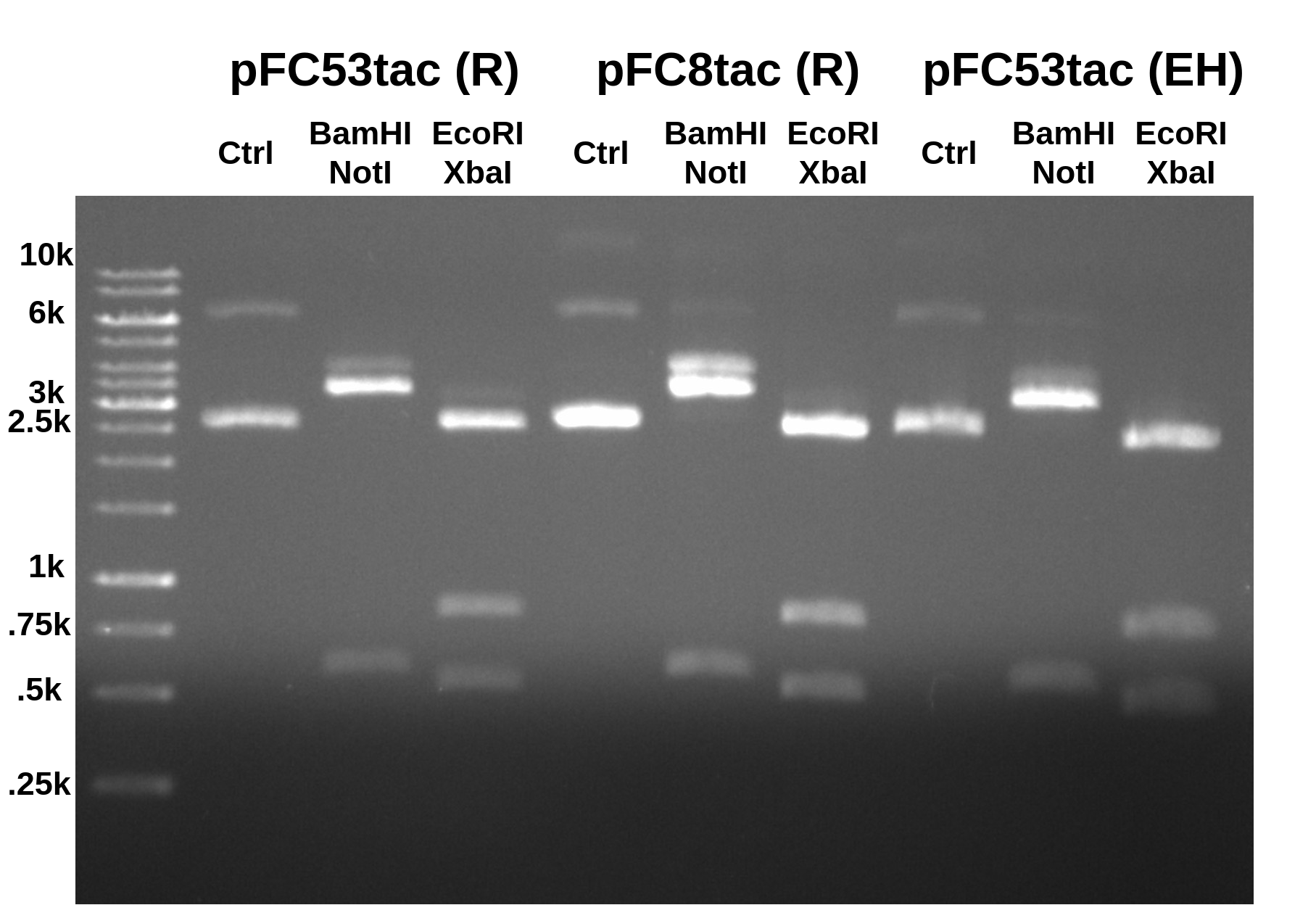
All digests look basically the same and match the expected results for the simulated digest of pFC8tac. At this point I would say with confidence that Rachel’s pFC53tac tube should be relabeled as pFC8tacT1T2. The double band produced by the EcoRI digest confirms the presence of the T1T2 terminators.
pFC primer design #
Primer design for sequencing pFC series plasmids. In order to be absolutely sure about the identity of each plasmid I will sequence each plasmid I am planning on working with using the primers in the below sections. This is my first time designing primers for the lab and so to keep track of them I created a spreadsheet were details of all future oligos should be stored.
pFC8tac #
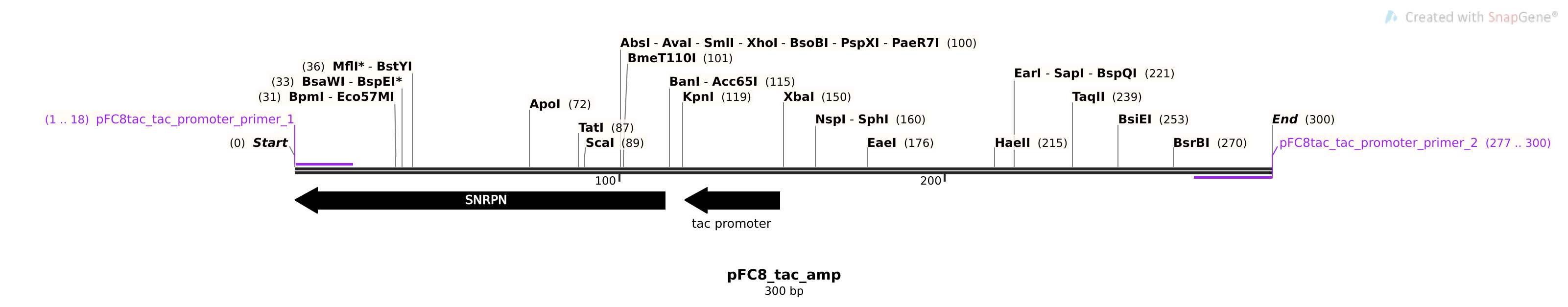
Sequences #
>pFC8tac_tac_promoter_Primer_1
gcagaacggcacaacagc
>pFC8tac_tac_promoter_Primer_2
gtattaccgcctttgagtgagctg
PCR program #
|95°C|95°C | |tmf:64.1
|____|_____ 72°C|72°C|tmr:66.4
|5min|30s \ 60.7°C _____|____|45s/kb
| | \______/ 0:30|5min|GC 55%
| | 30s | |301bp
pFC9 #
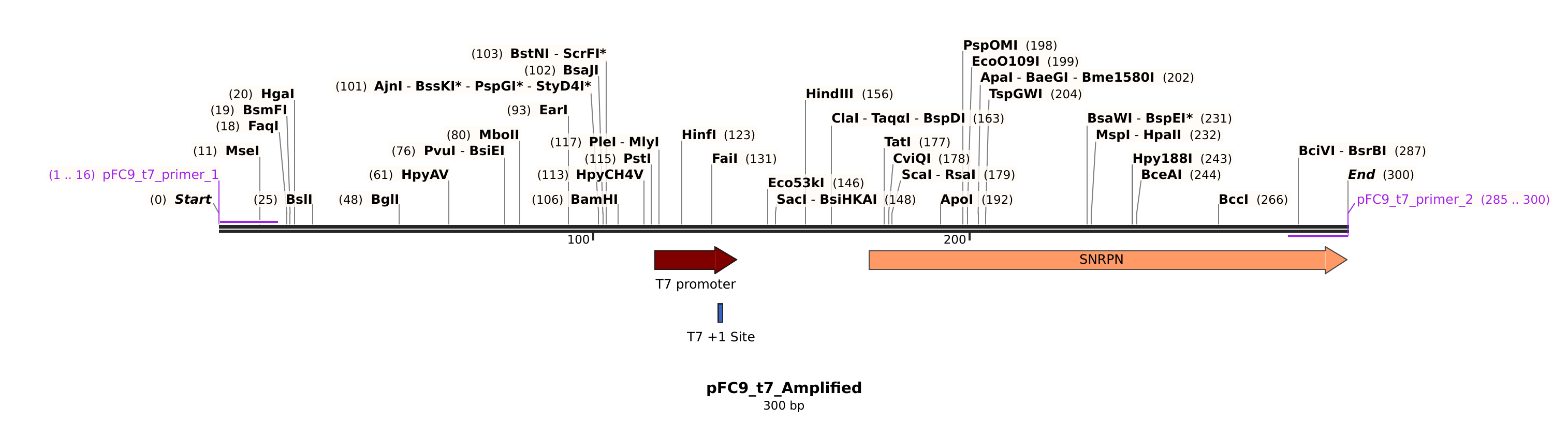
Sequences #
>pFC9_t7_primer_1
cccgccgcgcttaatg
>pFC9_t7_primer_2
gccccaatgcgagcgg
PCR program #
|95°C|95°C | |tmf:66.0
|____|_____ 72°C|72°C|tmr:69.1
|5min|30s \ 61.3°C _____|____|45s/kb
| | \______/ 0:30|5min|GC 56%
| | 30s | |300bp
pFC8 #
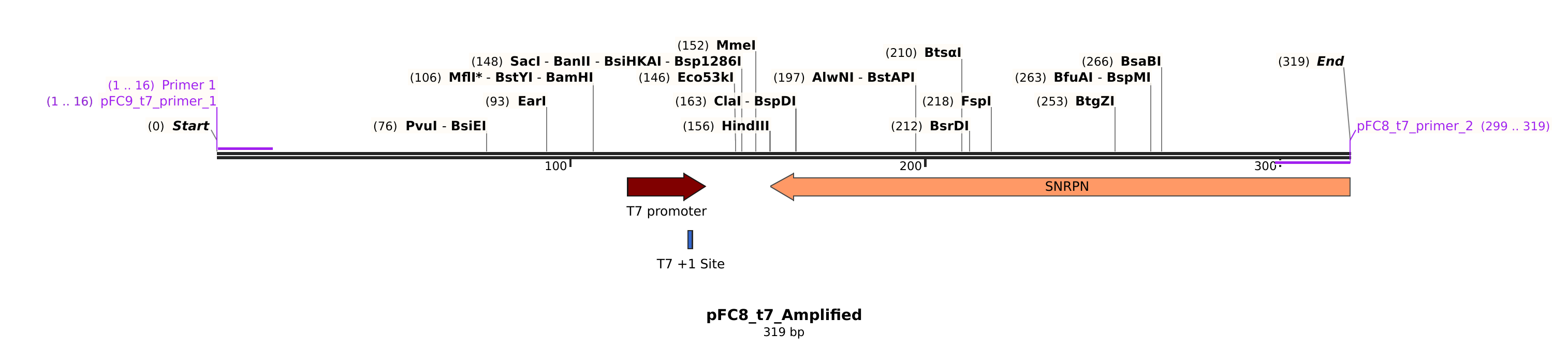
Sequences #
>pFC8_t7_primer_2
gaaggggcagtagcacagtcc
PCR program #
|95°C|95°C | |tmf:66.0
|____|_____ 72°C|72°C|tmr:69.5
|5min|30s \ 61.3°C _____|____|45s/kb
| | \______/ 0:30|5min|GC 55%
| | 30s | |319bp
All PCR programs calculated using pydna pcr module for Tac polymerase.
Notebook for protocol generation can be found here or pdf version here.
Insert sequence primers #
Since I was ordering primers I also added primers that will amplify all variable regions inserts in case we need to make a larger amount of any insert.
Sequences #
>Variable_region_insert_primer_1
tacgactcactatagggcgaattgg
>Variable_region_insert_primer_2
cctcctcgcctcggtcac
PCR protocol outline #
Reagent spreadsheet available at this link.