Confirming identity of pFC9(?) midi prep plasmid #
Today I am working on making sure pFC8 is 8 and pFC9 (or what I thought was pFC9) is also pFC8.
Double digest with BamHI and KpnI #
Replicating restriction digest protocol from yesterday to make sure that pFC9 is actually pFC8. Spreadsheet of reagents, volumes and DNA concentrations is available at this link.
Reagents #
| Reagent | Expiration | Lot number |
|---|---|---|
| CutSmart Buffer | 9/22 | 10046090 |
| KpnI-HF | 4/17 | 0061504 |
| BamHI-HF | 8/17 | 0101508 |
Expected results #
Simulated gel and lane contents are below.
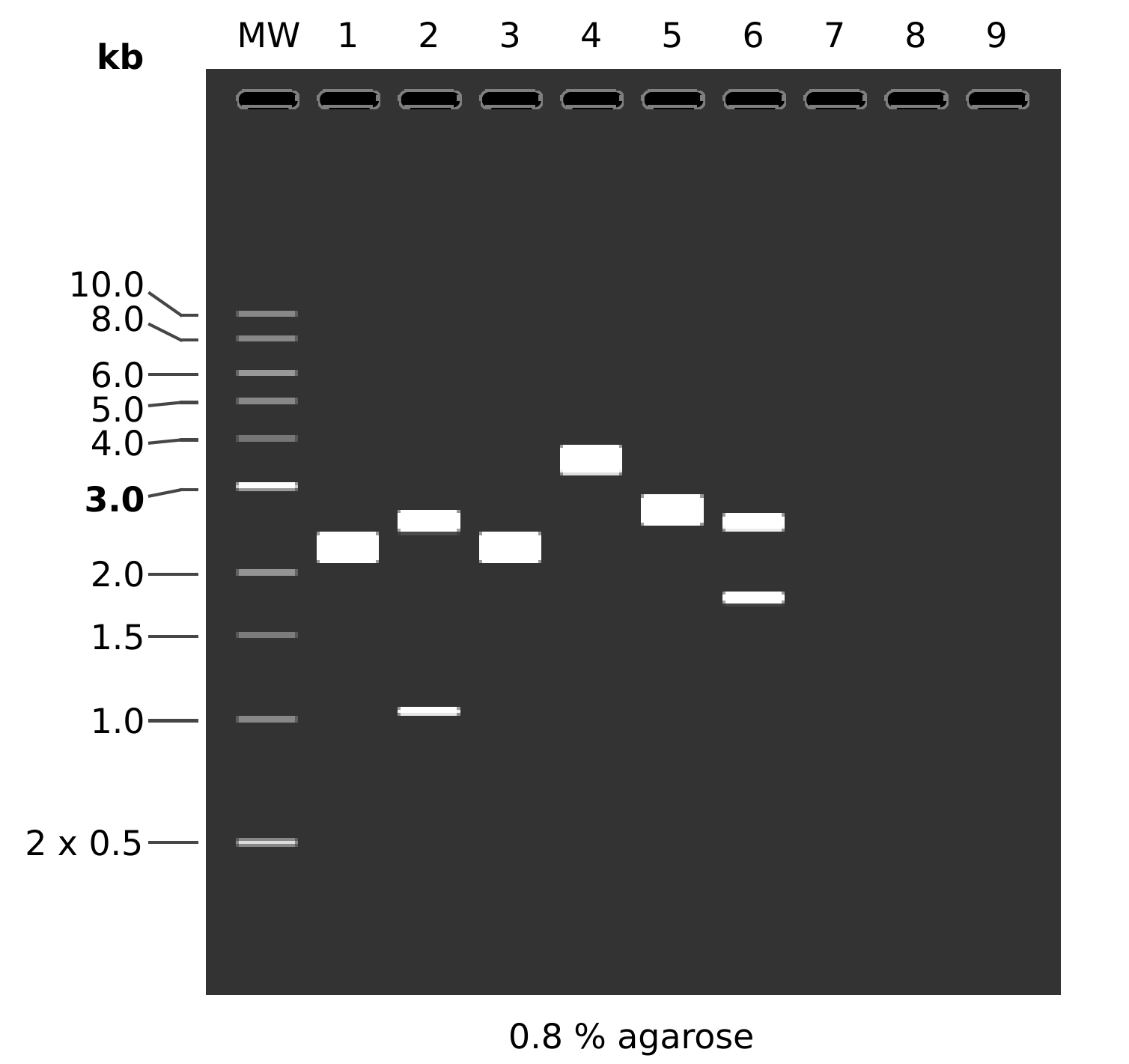
MW: 1 kb DNA Ladder
1: pFC8
1. 3589 bp
2: pFC8
BamHI + KpnI
1. 2558 bp
2. 1031 bp
3: pFC9
1. 3589 bp
4: pFC9
KpnI + BamHI
1. 3535 bp
2. 54 bp
5: pFC53tacT1T2
1. 4320 bp
6: pFC53tacT1T2
BamHI + KpnI
1. 2557 bp
2. 1763 bp
Results #
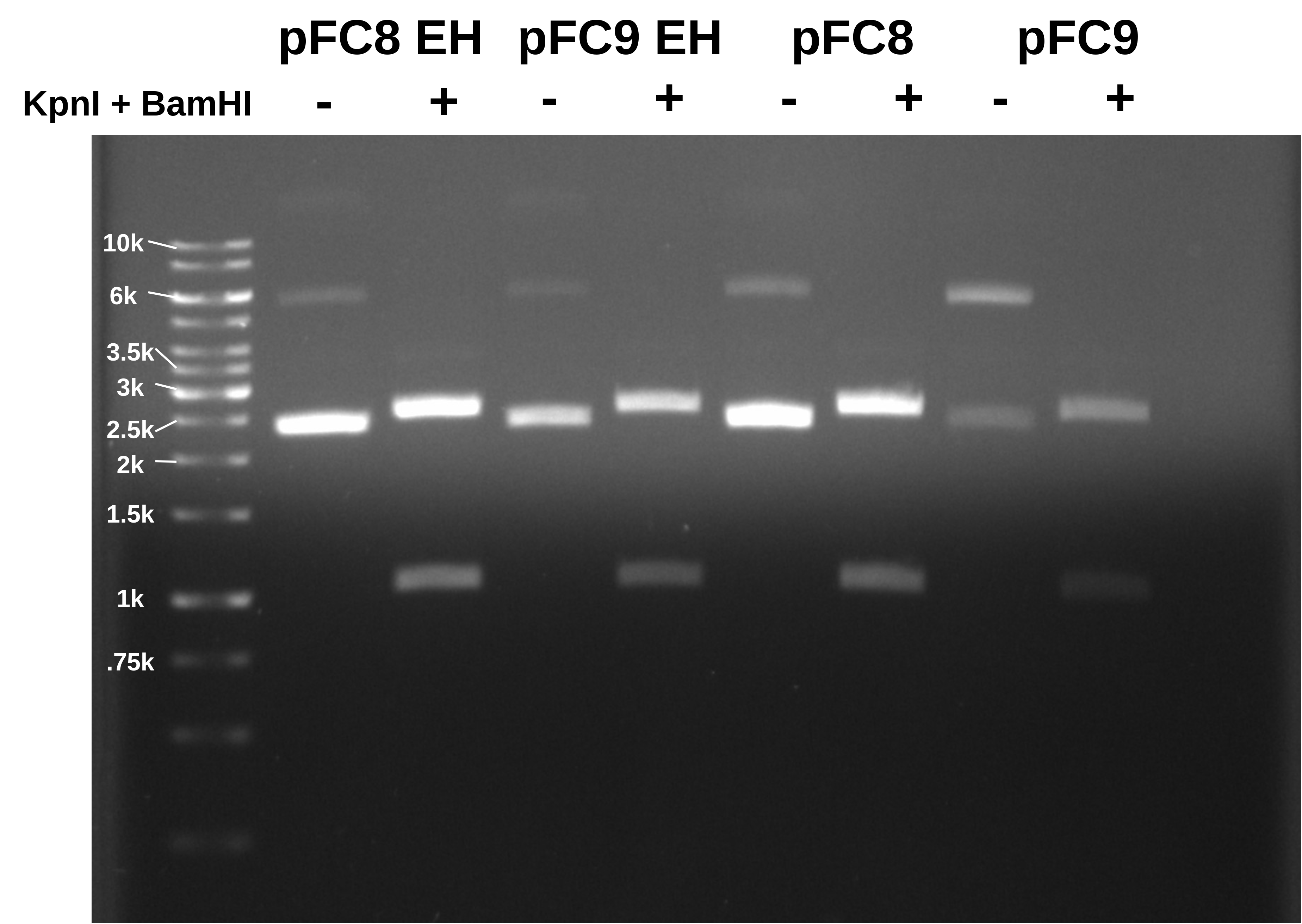
This looks like everything is pFC8!
Below is the tube I got “pFC9” from. I am pretty sure that says pFC9.

Talked to Fred after and we looked at the original primers and constructs used to make pFC8 and 9 and seems like the genbank file is incorrect. The locations of the KpnI and EcoRI sites are not flipped. This would explain the results of this digest. The pFC9 file has since been updated in the lab shared folder. I am currently reworking cloning strategy to reflect this. Will likely require new sequences or at least new homology arms.
Double digest with BamHI and NotI #
The R-loop forming region in pFC8 and 9 is flipped and NotI, as a part of this region, is also flipped with it. Here I am digesting pFC8 and 9 with BamHI (same location both plasmids) and NotI in order to tell them apart.
I followed exact same protocol as the BamHI + KpnI digest that is shown above but swapped KpnI-HF for NotI-HF. Should note that NotI-HF is very low and probably needs to be re-ordered.
Reagents #
| Reagent | Expiration | Lot number |
|---|---|---|
| CutSmart Buffer | 9/22 | 10046090 |
| NotI-HF | 12/20 | 10030791 |
| BamHI-HF | 8/17 | 0101508 |
Expected results #
Simulated digest is shown below. Only showing first four lanes as the next four should be the exact same in theory.
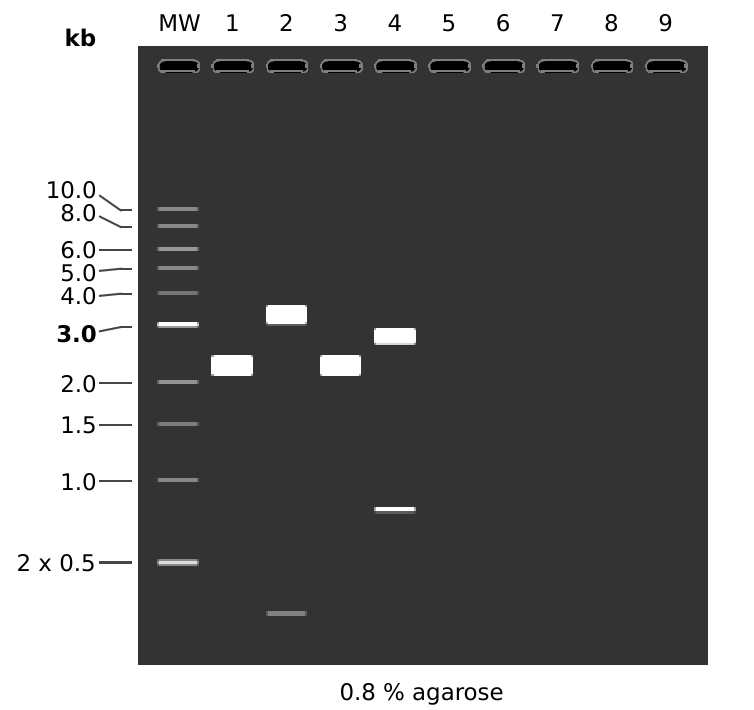
MW: 1 kb DNA Ladder
1: pFC8
1. 3589 bp
2: pFC8
BamHI + NotI
1. 3307 bp
2. 282 bp
3: pFC9
1. 3589 bp
4: pFC9
NotI + BamHI
1. 2801 bp
2. 788 bp
Results #
Ran gel 0.08 agarose for 1 hr at 120 volts with TAE and EtBr in running buffer as well as in agarose.
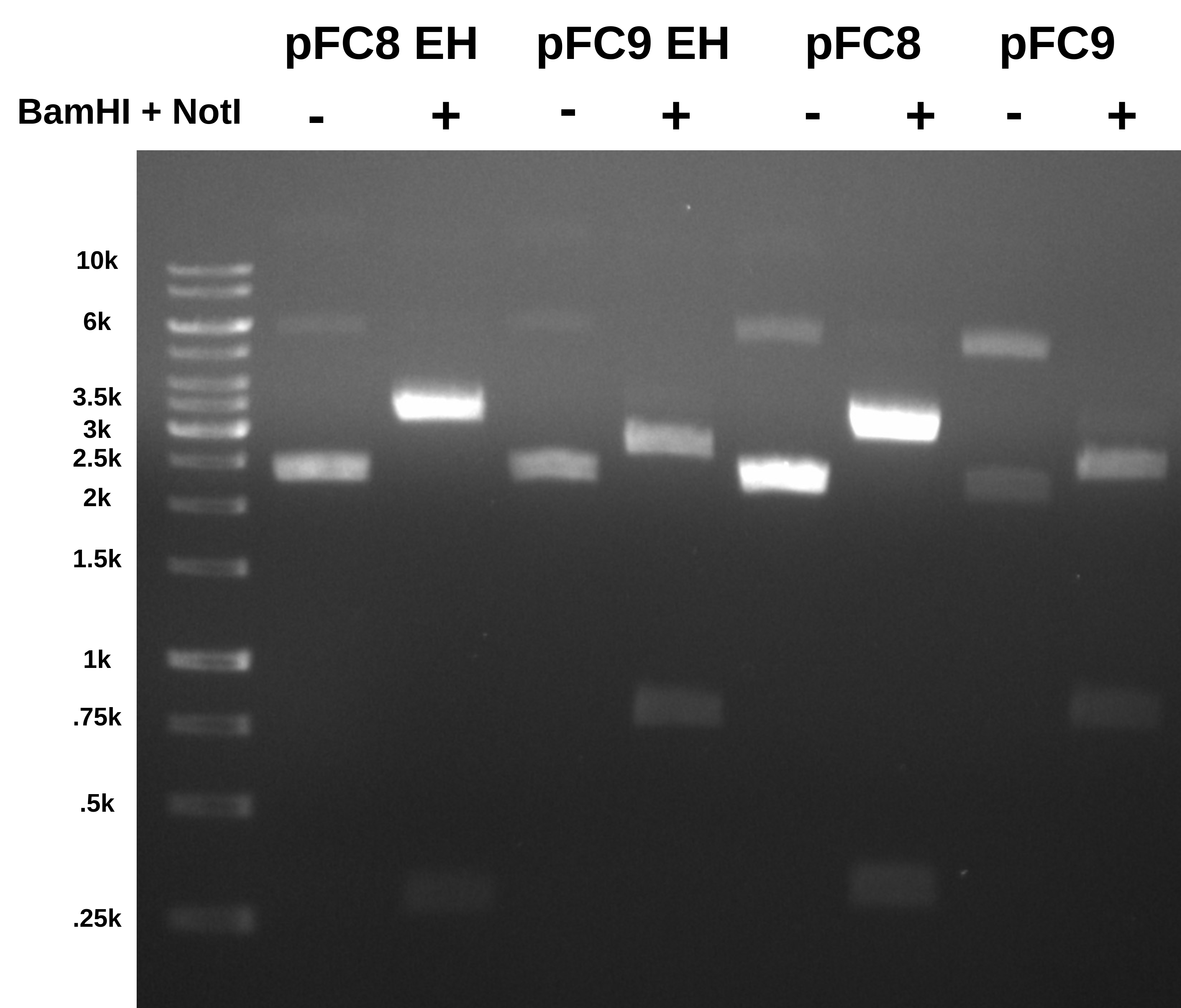
Based on these results pFC9 is not misclassified as pFC8 and is in fact correctly labeled.
Other misc. items #
pFC9 cultures #
Early in the day when I thought that my pFC9 midi prep sample was definitely pFC8 I started another pFC9 transformation. Those bacteria are currently plated and incubating in at 37C. I placed them into the incubation room around 9:45 am after electroporation with pFC9. I used the eletro-competent E.coli in the first box accessible in the -80C freezer.
Amp agar plates #
Made ~20 amplicilin agar petri plates for future transformations following the kitchen reagent sheet guidelines. I stored all plates in the plastic sleeve they came in in the deli fridge.
For tomorrow #
Tomorrow need to checkout bacteria and want to confirm the identity and orientation of the Tac promoter and T1T2 terminators using as many of the digests below that we have enzymes for.
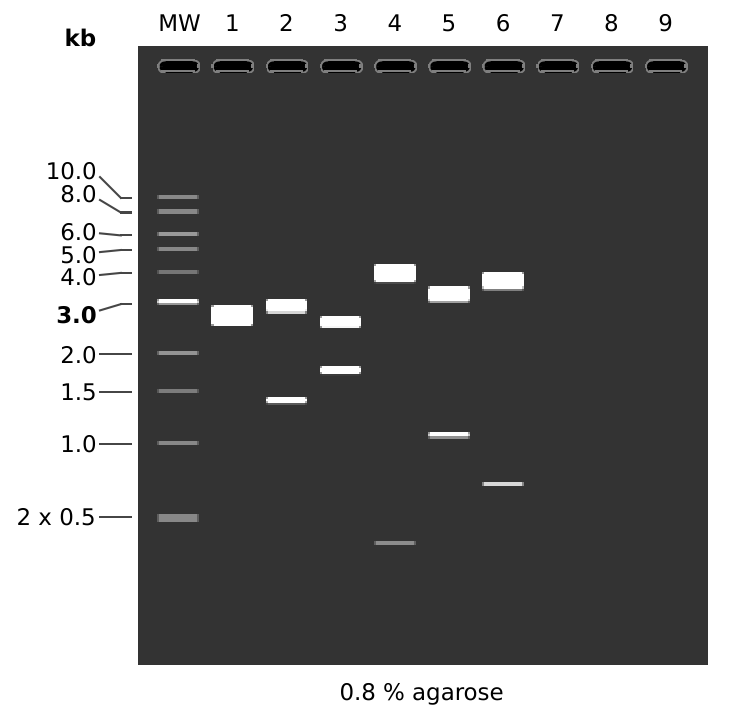
MW: 1 kb DNA Ladder
1: pFC53tacT1T2
1. 4320 bp
2: pFC53tacT1T2
HindIII + KpnI
1. 2935 bp
2. 1385 bp
3: pFC53tacT1T2
EcoRI + XbaI
1. 2574 bp
2. 1746 bp
4: pFC53tacT1T2
BamHI + HindIII
1. 3942 bp
2. 378 bp
5: pFC53tacT1T2
SacI + BamHI
1. 3258 bp
2. 1062 bp
6: pFC53tacT1T2
SacI + KpnI
1. 3619 bp
2. 701 bp
Lane 1 establishes undigested plasmid, second is a double check of the digest done on 7-26-21. Third digest is to partially confirm the presence of the T1T2 terminators since EcoRI is present in the terminators. Fourth digest confirms the length of the T1T2 terminators since this digest cuts that sequence out. Fifth and sixth digest together confirm distances between T1T2 and tac promoter since both cutting at the sacI site which is at the midpoint of the two features.