Confirming midi prep results #
Today I am working on visualizing the results of the midi preps I did for pFC8, 9 and 53tac (see notes from 7-20-21 to 7-23-21).
KpnI + HindIII double digest #
First I utilized a restriction digest with HindIII and KpnI to confirm plasmid ids and check for contaminating DNAs. Link to spreadsheet used for reagent calculation can be found here.
Reagents #
| Reagent | Expiration | Lot number |
|---|---|---|
| Buffer 2.1 | 5/23 | 10070034 |
| KpnI | 11/19 | 0551711 |
| HindIII | 8/22 | 10080749 |
KpnI was expired but it did not seem to significantly effect results.
Results #
I expected to see results similar to the simulated gel shown below.
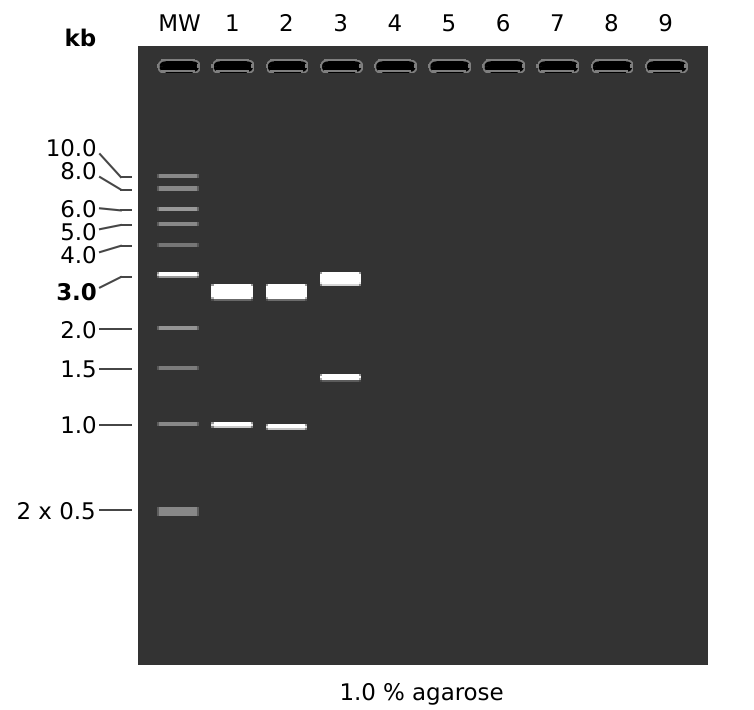
MW: 1 kb DNA Ladder
1: pFC8
HindIII + KpnI
1. 2608 bp
2. 981 bp
2: pFC9
HindIII + KpnI
1. 2616 bp
2. 973 bp
3: pFC53tacT1T2
HindIII + KpnI
1. 2935 bp
2. 1385 bb
I digested each plasmid for 30 mins at 37C and then ran them on a 0.8% agarose TAE gel for 1 hr at 90 volts.
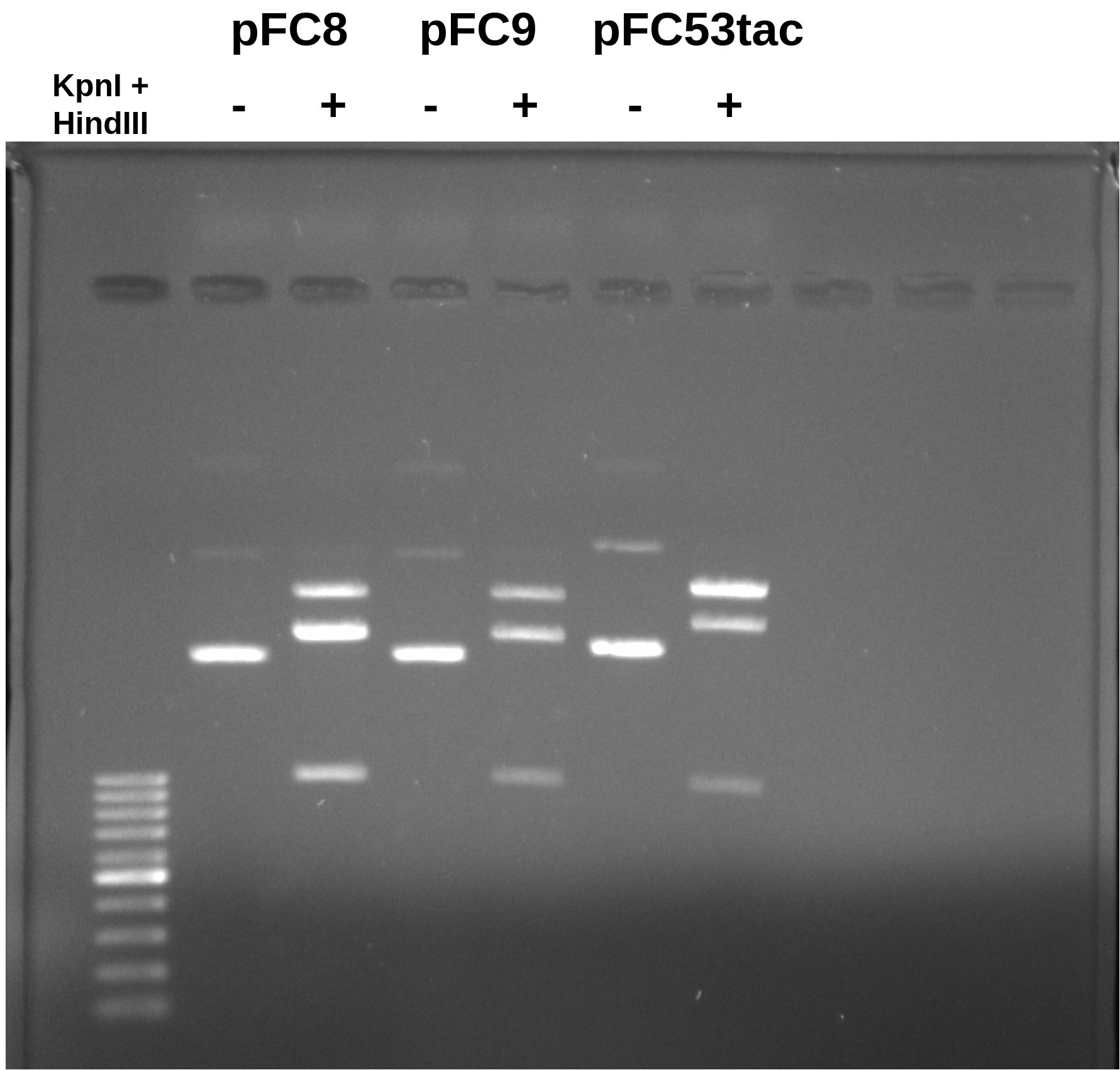
Overall both pFC8 and pFC9 look as expect although they cannot be distinguished with REs. pFC54tac does show the shift upwards on the large fragment but in a less dramatic fashion as the simulated gel. This is even more true for the smaller (expected 1385 bp) fragment which shows little differentiation from the ~980 bp fragments produced from pFC8 and pFC9.
BamHI-HF + KpnI-HF double digest #
After running the KpnI HindIII digest and looking back at the plasmid maps I realized there was a much better digest that I could have done that would in theory resolve all three plasmids from each other. Digesting with BamHI and KpnI takes advantage of the reversed position KpnI site on pFC8 and 9 and using BamHI which is located downstream of the T1T2 terminators on pFC53tac (which pFC8 and 9 lack) means all three plasmids will produce unique fragments.
So I repeated by protocol but used BamHI-HF and KpnI-HF enzymes with CutSmart buffer. I used HF version because these cutters normally do not have good activity together.
Reagents #
| Reagent | Expiration | Lot number |
|---|---|---|
| CutSmart Buffer | 9/22 | 10046090 |
| KpnI-HF | 4/17 | 0061504 |
| BamHI-HF | 8/17 | 0101508 |
Results #
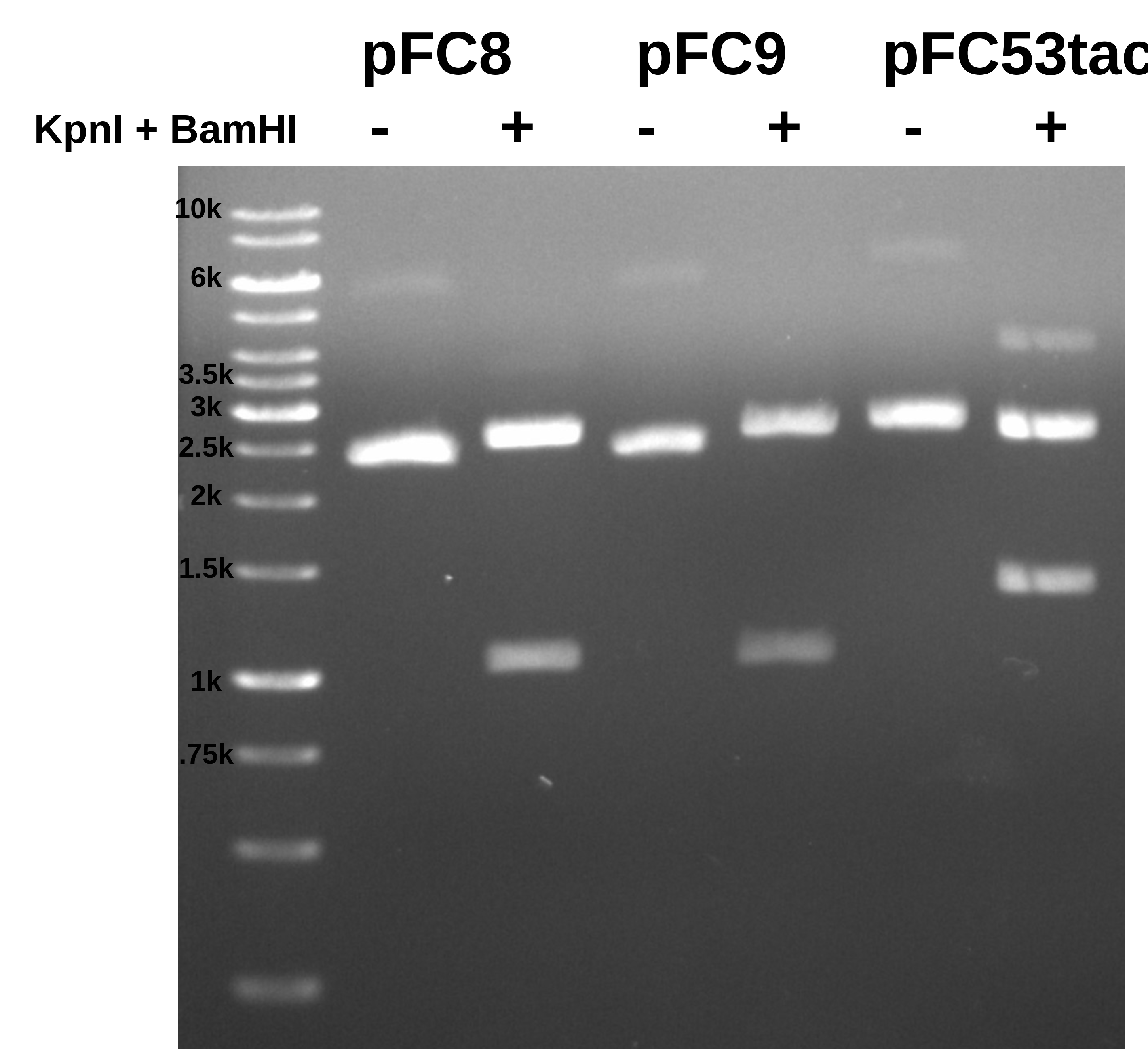
Here both pFC8 and pFC9 look like pFC8 in the simulated gel. This makes me think that I accidentally added pFC8 to the pFC9 tube of the transformation.
I think overall the results for pFC53tac conform to expectations, simulated gel for the two digests done below.
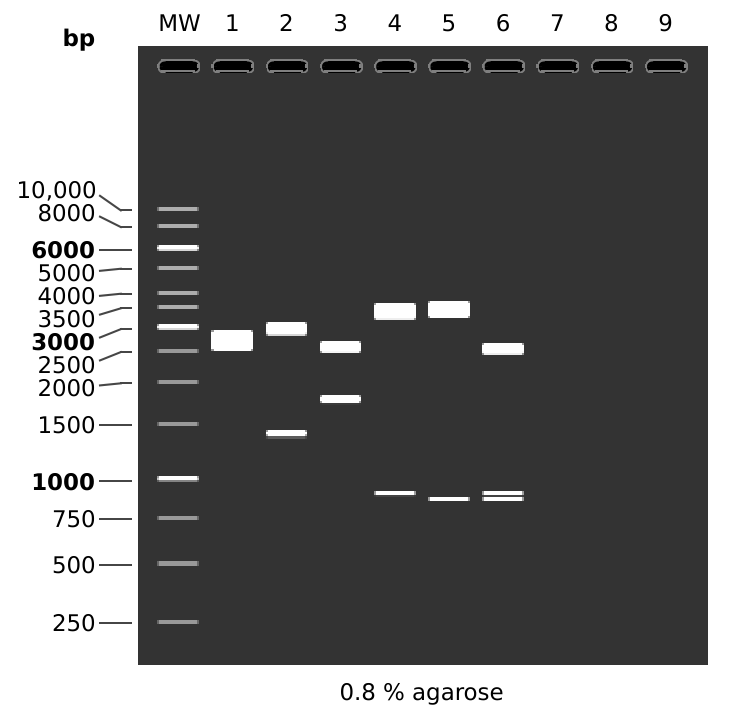
MW: 1 kb DNA Ladder
1: pFC53tacT1T2
KpnI + BamHI
1. 2557 bp
2. 1763 bp
2: pFC53tacT1T2
KpnI + HindIII
1. 2935 bp
2. 1385 bp
Tomorrow I will re-run this gel but using Fred’s pFC8 and pFC9 (the same plasmid used in notes from 7-20-21) and start transformation with pFC9 again because I am thinking that is the most likely explanation.