Gibson assembly reagent prep and VR fragment validation #
Gibson assembly reagent prep #
Prepped new Gibson assembly reagents; 5x isothermal buffer and 1.33x master mix according to the Gibson assembly recipe spreadsheet.
5x isothermal buffer #
| Reagent | Volume (ul) | Source |
|---|---|---|
| Tris HCL 1M | 1000 | Bench (8/23/21) |
| MgCl2 | 50 | Bench (8/19/21) |
| PEG 8000 80% | 625 | Made 9/4/21 |
| NAD 50 mM | 200 | Made 9/4/21 1ml aliquote |
| DTT | 1 | PCR box |
| DNTPs 10 mM | 20 | Made 9/29/21 |
| npH20 | 104 | Fresh Dnase Rnase free PCR grade |
Gibson 1.33x master mix #
| Reagent | Volume (ul) | Source |
|---|---|---|
| Tris HCL 1M | 1000 | Bench (8/23/21) |
| Reagent | Volume (ul) | Source |
| Taq ligase | 13.25 | NEB |
| 5x isothermal buffer | 20 | 2ml fraction made 9/4/21 |
| Taq DNA Pol | 0.6 | NEB purchased 9/29/21 |
| T5 Exo | 0.1 | NEB |
| H20 | 60 | Fresh Dnase Rnase free PCR grade |
BglII digestion followed by PCR product validation #
Testing that BglII digestion followed by PCR amplification from 10/1/21 are the correct lengths by using VR-31 PCR product as a loading control. VR-31 was was supplied directly as a fragment and so PCR product should be exactly the correct length. All samples were composed of the reagents below.
- 3 ul PCR product
- 8 ul npH20
- 3 ul purple loading dye
Loaded all samples onto 0.8% 1x TAE agarose gel with 2 ul EtBr added to both the gel and 20 ul EtBr in the 1x TAE running buffer. Lane layout is shown in the table below.
| Lane | Sample | Lane | Sample |
|---|---|---|---|
| 1 | 1 kb ladder | 6 | VR-16 |
| 2 | VR-31 | 7 | VR-17 |
| 3 | VR-8 | 8 | VR-20 |
| 4 | VR-12 | 9 | VR-21 |
| 5 | VR-15 |
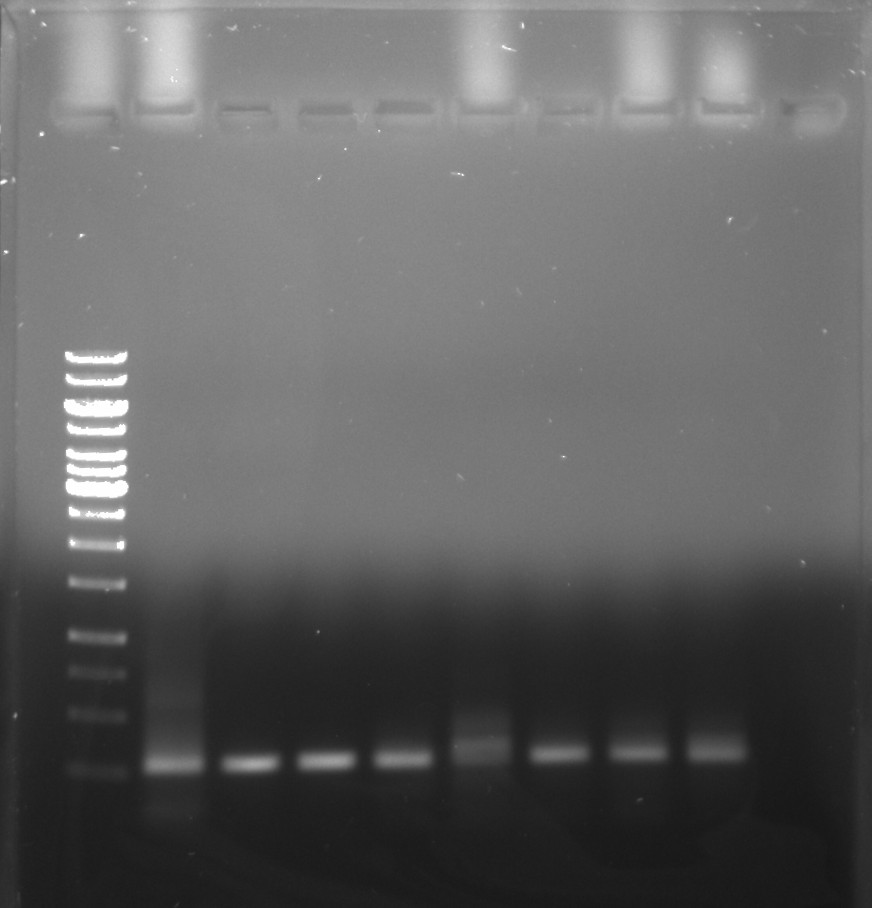
Overall bands look like they are in the right place and so I moved on with the agarose gel extraction.
BglII digestion followed by PCR AGE #
Started gel with all BglII -> PCR samples for agarose gel extraction. Made the gel with 100 ml 0.8 agarose with 4 ul EtBr using 1x TAE for both the agarose gel and the running buffer. Added 40 ul EtBr to the running buffer. Ran gel at 120 V for 45 mins on undergrad bench. After running gel extracted VR insert bands using tin foil method on the trans-illuminator. Extracted DNA from bands using freeze and squeeze protocol.
Agarose extraction yields #
| Sample | ng/ul | Sample | ng/ul |
|---|---|---|---|
| 8 | 281.9 | 21 | 420 |
| 12 | 268 | 23 | 433 |
| 15 | 474 | 25 | 297 |
| 16 | 314 | 27 | 205 |
| 17 | 360 | 31 | 311 |
| 20 | 342 |
Agarose gel extraction product gel #
Given the high yield of the extractions I went ahead and ran the extracted products out on another gel. I loaded ~500 ng of each sample along with 3 ul purple loading dye and 8 ul H20 onto 0.8% 1x TAE agarose gel. Run the gel for 45 mins at 120V. Lane layout is below.
| Lane | Sample | Lane | Sample |
|---|---|---|---|
| 1 | 30 | 7 | 20 |
| 2 | 8 | 8 | 21 |
| 3 | 12 | 9 | 23 |
| 4 | 15 | 10 | 25 |
| 5 | 16 | 11 | 27 |
| 6 | 17 | 12 | 31 |
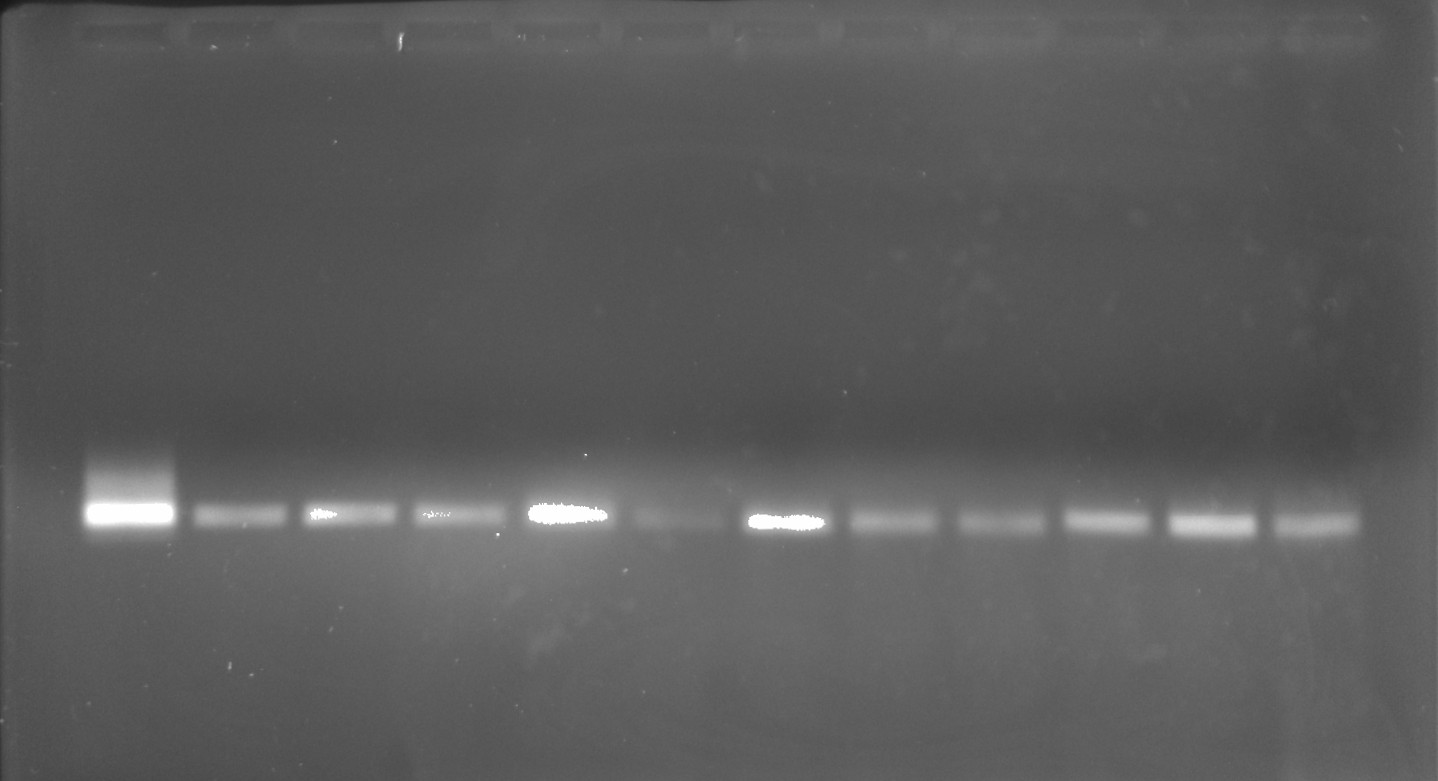
All bands look clean and are at the correct height (VR-30) loading control in lane 1. Moving forward with Gibson assembly of these fragments into pFC9.