Chloroquine gel post staining and imaging #
Picked up gels that I started yesterday. Washed each in ~500 ul of npH20 for two hours with shaking. After I drained H20 and poststained with 200 ul 1 ul / ml EtBr for 45 minutes. After post stained rinsed for 5 mins with EtBr.
Gel layout #
| Gel | Lane | Treatment |
|---|---|---|
| 1 | 1 | Empty |
| 1 | 2 | Control |
| 1 | 3 | Topo |
| 1 | 4 | Topo + Gyrase |
| 2 | 1 | Empty |
| 2 | 2 | Control |
| 2 | 3 | Topo |
| 2 | 4 | Topo + Gyrase |
Gel images #
DNA is not present on the gel. Looking back at my calculations I used way too much chloroquine in the gel. Maybe this caused bands to run backwards?
Made stock solution of chloroquine with 125 ug / ul and prepared new Gyrase activity assay as described in 9/27/21 notes and prepared chloroquine gel that actually follows the correct protocol using 2 ml of stock chloroquine solution in 98 ul agarose. Loaded samples and ran gel at 110V for 16 hours.
PCR contamination test #
Testing new PCR master mix with various Taqs. Using reagents I have never used before to test for Taq contamination (in theory).
Master mix #
| Reagent | Lot |
|---|---|
| 5x GoTaq | 28522702 |
| NEB dNTP | 0561010 |
| H20 | NA Fresh |
Polymerases #
| Pol name | Lot |
|---|---|
| Phusion Pol | 00783326 |
| Deepvent | 11 |
| Lab taq | NA (A on tube) |
Made four reactions one with each polymerase and one negative control with only master mix. Amplfied all samples for 34 cycles using standard PCR protocol.
Gel #
Ran products on 0.8% agarose TAE gel with 1ul/ml EtBr in 1x TAE buffer at 120V for 45 mins.
| Lane | Sample |
|---|---|
| 1 | MM + Phu |
| 2 | MM + Deepvent |
| 3 | MM + lab taq |
| 4 | MM |
| 5 | 1kb ladder |
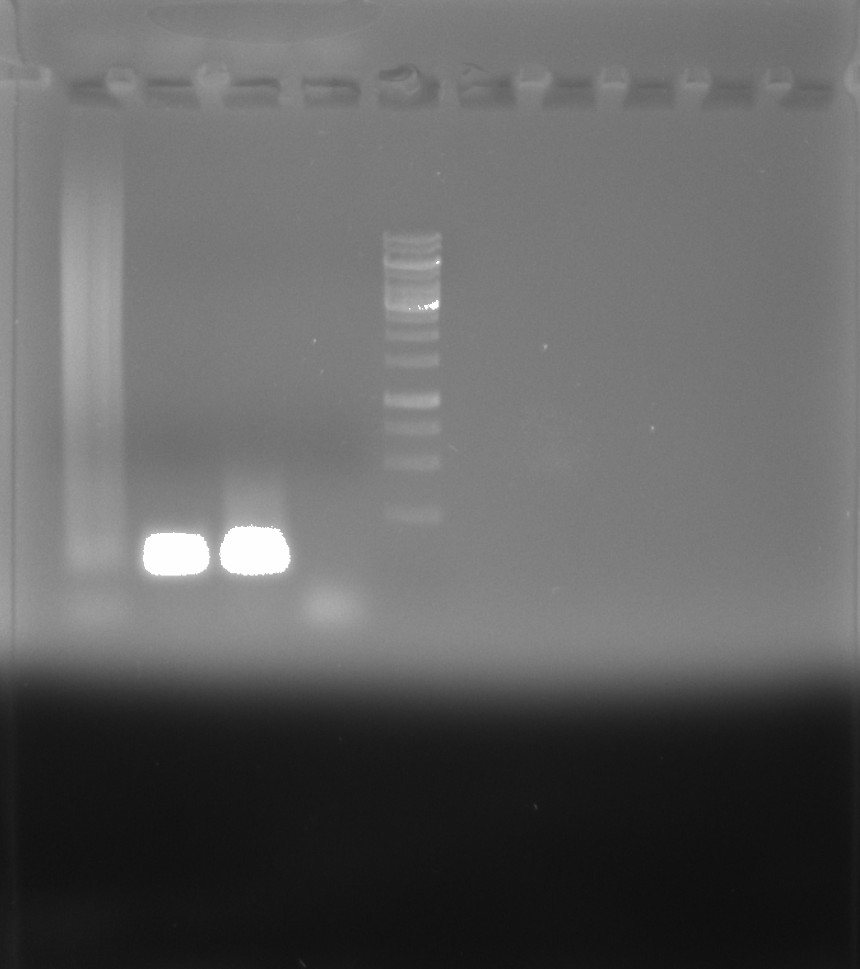
Clearly contamination present in the Deepvent (lane 2) and lab taq (lane 3) lanes. Not sure what is going on with Phu that is producing a smear but Megan saw this in an unrelated test that used phusion but with no substrate (as is the condition here).
Need to get even more extreme in contamination battle.