pFC9VR28 LiCl purification recovery #
Finally got around to precipitating DNA from the somewhat botched LiCl prepcipitation where I used 70% EtOH instead of 100%. Added 1/10th volume of sodium acetetate, froze at -80C for 1 hour and then spun at 10k RPM (max large lab centrifuge) for 1.5 hours, washed with 70% EtOH, then dried pellet and resuspended in 2ml 10mM Tris HCL. Final concentration was 40 ng/ul. Little lower than hoped but at least got the DNA out.
PCR and BglII digestion product cleanup #
Yesterday I worked on making Gibson ready fragments from inserts cloned in vectors. I PCRed inserts first, then digested with BglII see yesterday’s notes. I then did a EtOH precipitation on all fragments following the protocol below, although this was only strictly necessary on BglII digested samples to remove BglII.
- Add 1/10 volume 3M ph 5.2 sodium acetate
- Add 2.2 volumes 100% ethanol
- Freeze at -80 for 1 hour
- Spin at max speed at 4C for 30 minutes
- Decant, but save, the supernatant
- Wash pellet with 70% EtOH
- Spin for 5 mins at max speed
- Decant supernatant
- Dry pellet for 15 mins at room temperature
- Resuspend in 40 ml 10 mM Tris HCL
Following this protocol I then OD each sample results of which are below.
| Sample | ng/ul |
|---|---|
| 6 | 18.2 |
| 7 | 23.6 |
| 8 | 11.4 |
| 9 | 21.5 |
| 12 | 12 |
| 15 | 9.7 |
| 16 | 16 |
| 21 | 4.3 |
| 23 | 23 |
| 26 | 10.9 |
| 27 | 27 |
| 28 | 10.6 |
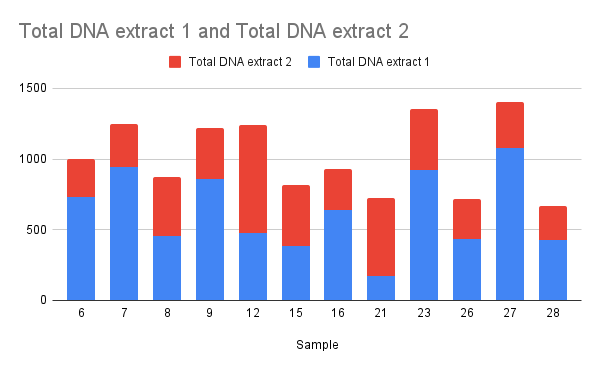
Yields seemed a bit low to me, with 40ul of sample this represented about 50% recovery for most samples. So I repeated the protocol on the supernatent to see if I could get any extra DNA; OD results are below.
| Sample | ng/ul |
|---|---|
| 6 | 13.7 |
| 7 | 15.3 |
| 8 | 20.9 |
| 9 | 17.9 |
| 12 | 38.1 |
| 15 | 21.4 |
| 16 | 14.5 |
| 21 | 27.5 |
| 23 | 21.8 |
| 26 | 14 |
| 27 | 16.1 |
| 28 | 12.1 |
This recovered a significant amount of DNA and got efficiency up to around 70%. I did not combine samples and stored in the deli fridge.
Then I ran ~200 ng of each fragment out on a 0.8 gel at 120V for 45 mins.
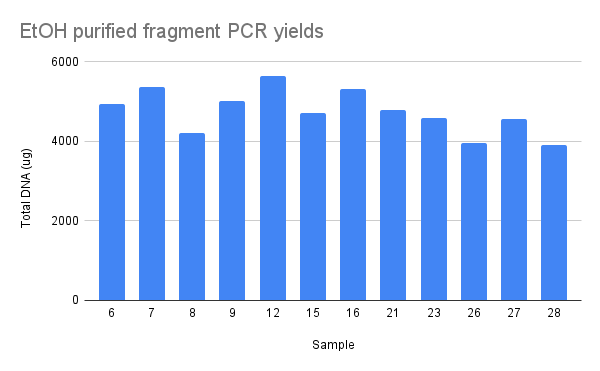
I then used ~10ng of each in a PCR reaction in order to create highly concentrated fragment samples. I used a standard PCR master mix with lab taq and OneTaq NEB buffer. OD of PCR products is chart below.
BglII digest followed by PCR #
Based on the results of the gel it looks like the digestion may be incomplete. So I set up the reverse of the reaction that produced these fragments. First digesting ~200 ng of insert containing vector for 1 hour and then adding PCR master mix directly to sample and amplifying whatever is in there (hopefully the digested fragment).
| Reagent | Volume (ul) |
|---|---|
| BglII | 0.2 |
| NEB 3.1 10x buffer | 1 |
| DNA | 2 |
| H20 | 2.4 |
Ran this reaction for 1hr @ 37C in the thermocycler and then added 25 ul PCR master mix (lab taq, NEB OneTaq buffer) and ran reaction overnight before leaving the lab.
