Insert fragment synthesis continued, BglII digestions and revisiting IVT #
Transformation efficiency of old chemically competent cells #
Yesterday I set up a control transformation to measure the efficiently of old chemically competent cells I found in the -80C. Three colonies grew equating to an efficiency of 1.9e6 transformants / ug plasmid. On the low end of what is acceptable but potentially still workable.
Insert fragment creation continued #
Overnight PCR results #
Yesterday I set a PCR to run as I was leaving the lab. OD results from each sample are in the table below.
| Insert | ng/ul |
|---|---|
| 6 | 114.6 |
| 7 | 123.6 |
| 8 | 129 |
| 9 | 87 |
| 12 | 123 |
| 15 | 109 |
| 16 | 151 |
| 21 | 66 |
| 23 | 342 |
| 26 | 340 |
| 27 | 303 |
| 28 | 220.9 |
Now we have about double the DNA. Turns out its important to have nucleotides in your PCR mix. Although there must have been some free from the thermo synthesis reactions because even without adding there was some degree of amplification. Either way there was enough DNA to move onto BglII digestion.
Digestion of successful PCR reactions #
Next, having enough DNA to move forward I digested vector originating samples (8, 12, 15, 16, 21, 23, 26, 27, 28) with BglII using the reagents and volumes in the table below.
| Reagent | Volume (ul) |
|---|---|
| PCR product | ~25 |
| BglII | 1 |
| r3.1 10x buffer | 5 |
| H20 | 19 |
Results after 1.5 hours #
OD samples after digestion shown in the table below.
| Sample | ng/ul |
|---|---|
| 8 | 51 |
| 12 | 49.9 |
| 15 | 4.6 |
| 16 | 66.5 |
| 21 | 28.3 |
| 23 | 52.7 |
| 26 | 50.8 |
| 27 | 32.8 |
| 28 | 42.5 |
Overall looks as expected considering the dilution that occurs by adding the digestion reagents except for VR-15 which lost most DNA apparently. Not sure where it went though. Next, I ran BglII digested samples on 0.8 agarose gel for 40 mins at 120V using ~200 ng of DNA. The gel showed underdigestion of a number of fragments. The lane layout is shown in the table below since I do not want to have to label this one. First lanes of each row were inserts amplified from fragments so if digestion is complete, all bands should be at this height.
| Lane | Sample | Frag? |
|---|---|---|
| 2 | 6 | 1 |
| 3 | 7 | 1 |
| 4 | 8 | 0 |
| 5 | 12 | 0 |
| 6 | 15 | 0 |
| 7 | 16 | 0 |
| 12 | 9 | 1 |
| 13 | 21 | 0 |
| 14 | 28 | 0 |
| 15 | 26 | 0 |
| 16 | 27 | 0 |
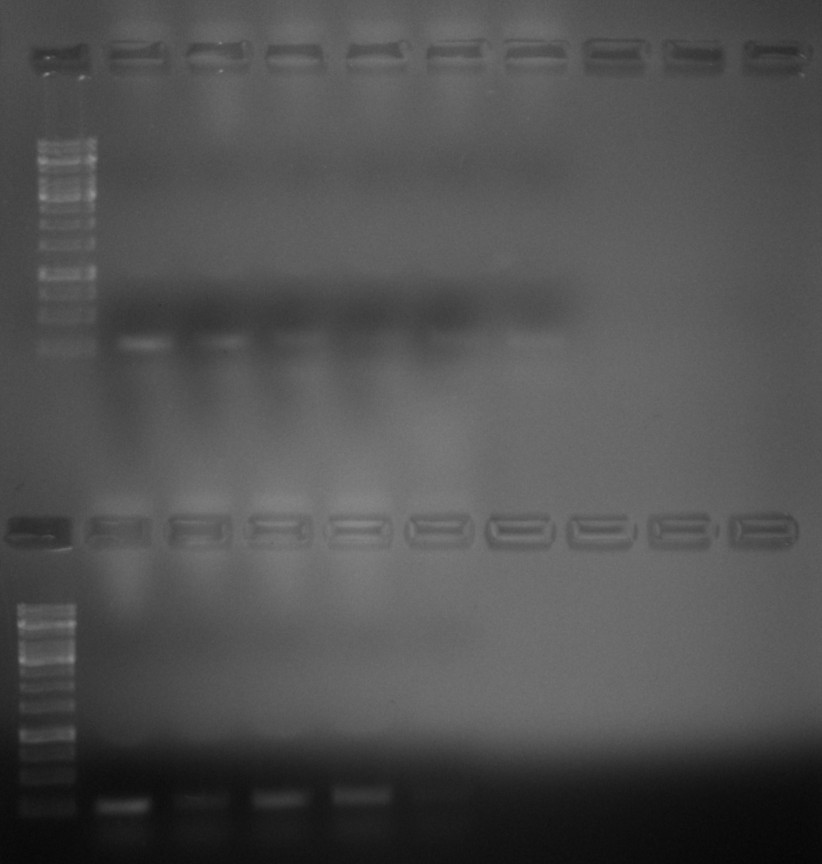
So I put BglII digestions back into the 37C room to sit for a while while I revisited IVT reactions. After about 3 hours digesting at 37C I reran the gel using ~200 ng of each sample at 120V for 45 mins in TAE with EtBr.
Bands are all about at the same height here. Lane 2 would be the control to compare digested fragments to. I put gel into fridge to cool faster but it was not level causing some lanes to have slightly thicker agarose than others which could explain the sort of curved shape. Despite this there is much less difference in height to the 1.5 hour digestion so I will move forward with these fragments.
IVT pFC53, pFC9 and pFC8VR13 #
Attempt 1 #
Followed lab protocol for IVT reactions with pFC53, pFC9 and pFC8VR13. Design for each reaction is described in this spreadsheet table 9-14-21 and shown in the table below (these are main mixes and do not include RNAseA or H addition steps).
| Plasmid | DNA (ul) | 5x Transcription buffer (ul) | DTT | rNTP (ul) | DTT (ul) | npH20 (ul) | Pol Species |
|---|---|---|---|---|---|---|---|
| pFC53 | 1.2 | 6 | 3 | 0.6 | 3 | 16.2 | T3 |
| pFC9 | 2 | 6 | 3 | 0.6 | 3 | 15.4 | T7 |
| pFC9VRX | 1 | 6 | 3 | 0.6 | 3 | 16.4 | T7 |
After completion of each reaction, I ran products out on 0.8 agarose TBE gel at 60V for 2 hours with purple loading dye. At this point I realized I forgot to add DTT to the main mixes (oops forgetting reagents always seems to be my downfall) but the gel had been run.
It appears that pFC53 was not transcribed really at all (lack of RNA smear at the bottom of the gel) otherwise this looks really great! Efficiency of pFC9 and pFC9VR13 are around 95% by my estimate which is far better than any IVT reaction I had done previously. In next attempt I will be sure to add DTT. If pFC53 forms R-loops then it must be that T3 is sensitive to the presence of DTT while T7 is not so much.
Attempt 2 #
This time around I made sure to add DTT. Additionally loaded the gel using the lab reciepe for SDS loading dye. Gel was ran at 60V for two hours same as the last one. 0.8 agarose in TBE. Both gels were post-stained for ~45 mins in EtBr.
Here the shift in pFC53 is much clearer, indicating that T3 is in fact sensitive to DTT. Using the SDS loading dye does not appear to make that much of a difference.


