Gibson prep and sanger analysis #
Sanger sequence analysis #
On Wendesday I submited two batches of samples for sequencing; both from Gibson assembly reactions. The first was from reactions completed on 9/5/21 and the second from reactions completed on 9/8/21.
9/5/21 Gibson assembly Sanger analysis #
First I used BLAST to align the VR insert reference sequences to the submitted samples (format VR-{insert number}-{colony number}). Since BLAST does not align low similarity sequences I created the second plot to show which Sanger sequences had no alignment.
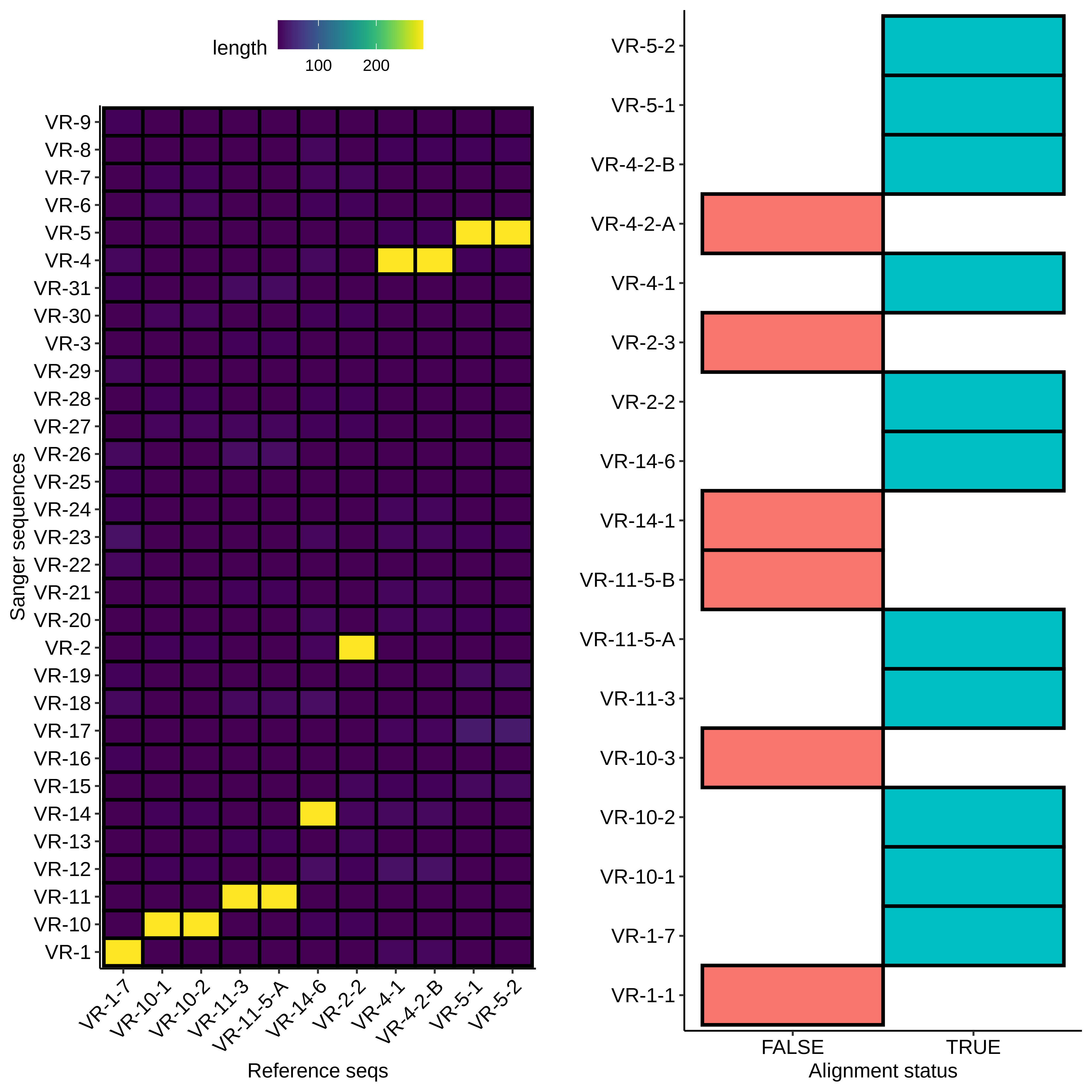
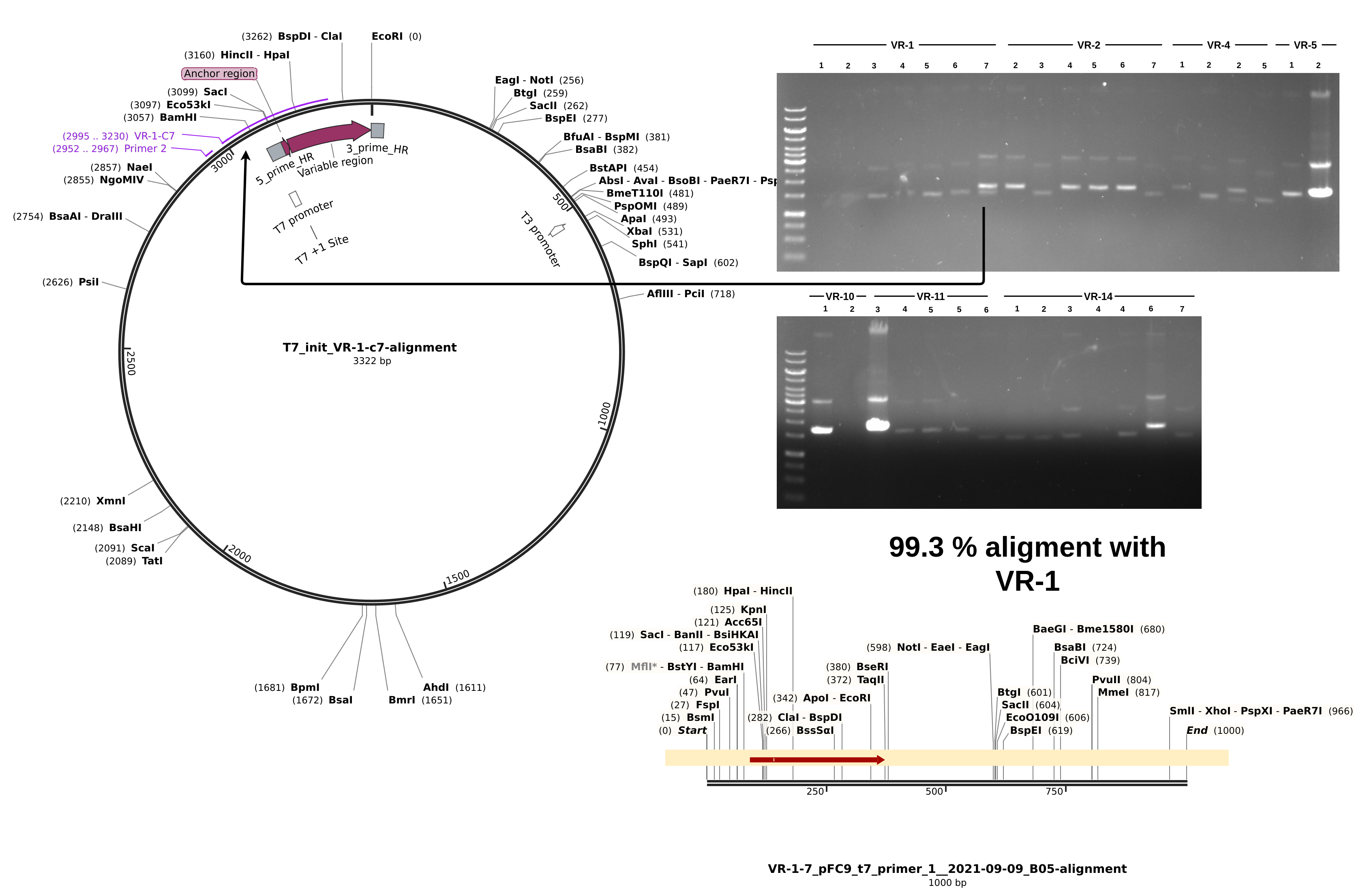
For sequences that did align, everything looks about as expected. Longest alignment lengths map to the insert each sample is labeled as. If we look at a non-alignment, for example VR-1-1, we can see a couple of interesting things.
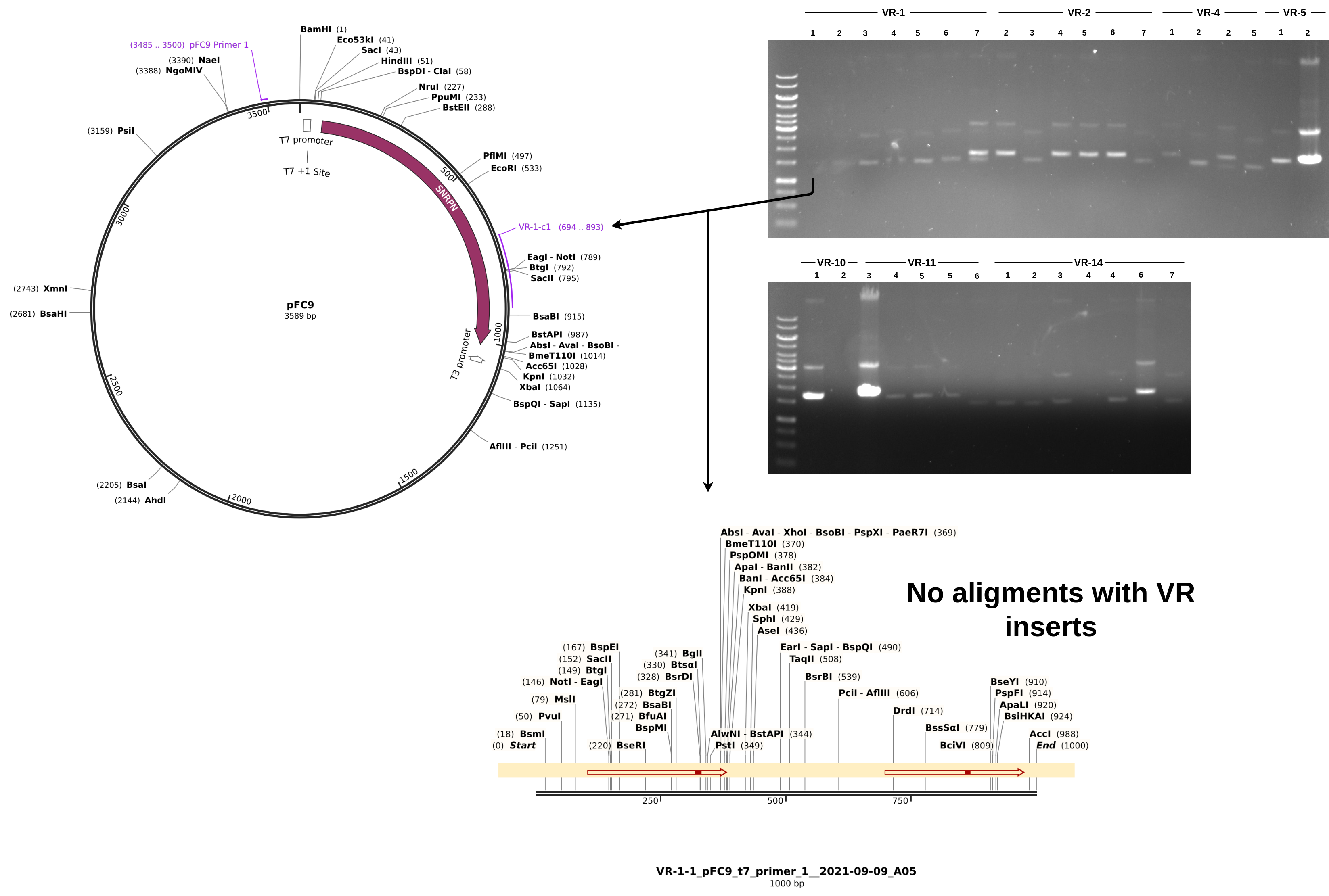
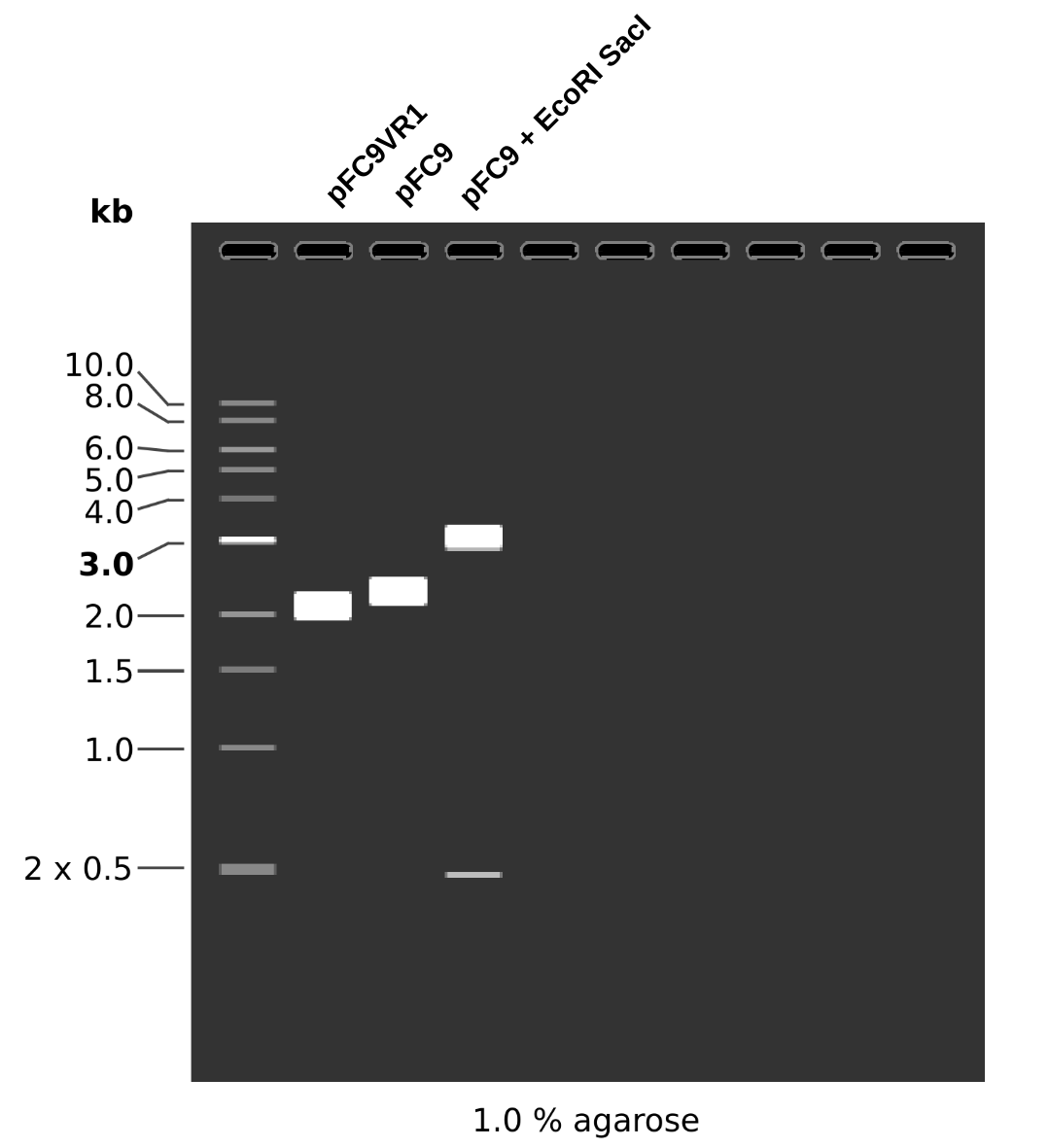
Although the Vr-1-1 band is faint it is there and it about at the same height as samples VR-1-1 through VR-1-6. From the simulated gel below we can see that plasmids with a successful insertion will run slightly but detectably lower on the gel due to their smaller size compared to pFC9.
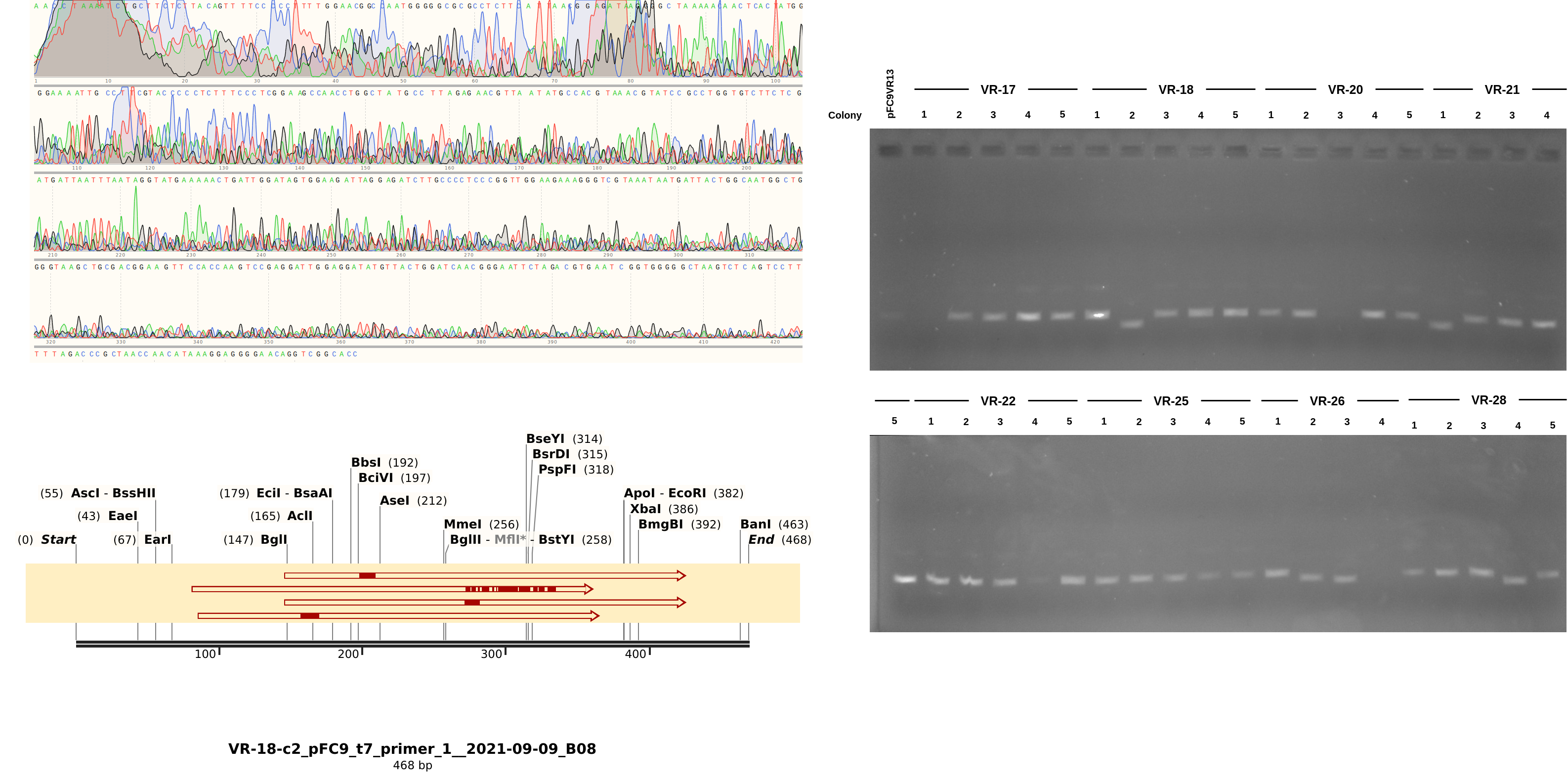
9/8/21 Gibson assembly Sanger analysis #
With the fact that the height of midi-prep products on gels can be an early indicator of a likely successful insert (or at least disqualify potential ones) we can take a look at the results from the 9/8/21 Gibson assembly. The heatmap type plots of BLAST sequence alignments are shown below.
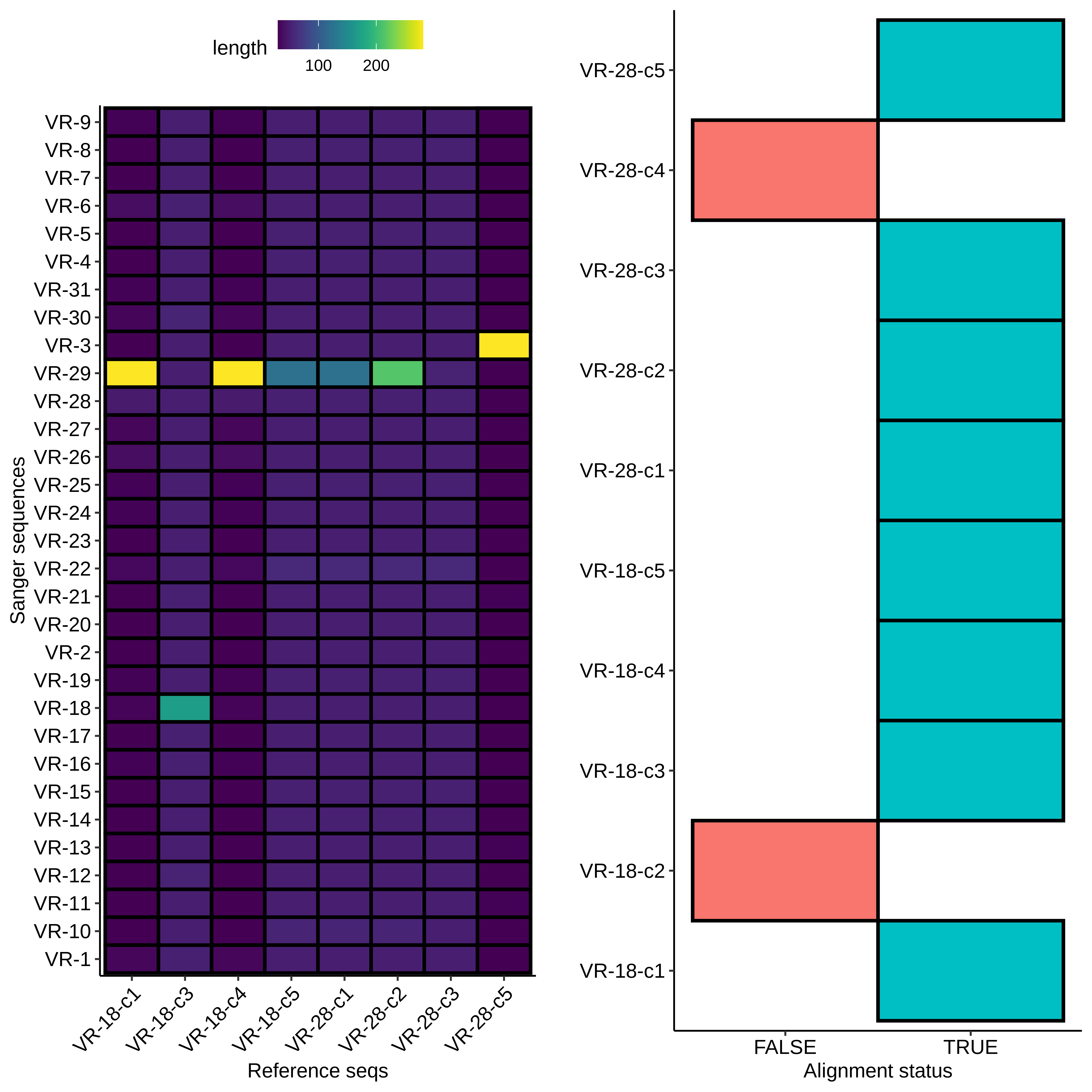
Here the results are a bit more muddled. Most of the sequences did align to a reference to some degree but almost none aligned to their labled reference. THis could be explained by sample mix up but I really focused on sample organization this time around and VR-29 nor VR-3 were ever actually a part (at least to my knowledge) of this round of Gibson reactions. Either way, looking more closely at the data and traces yields a few pieces of information. First looking at a “successful” alignment (in so much as an insert made it into the pFC9 fragment), VR-18-1 showed all the signed of successful insertion but with fragment VR-3 not 18.
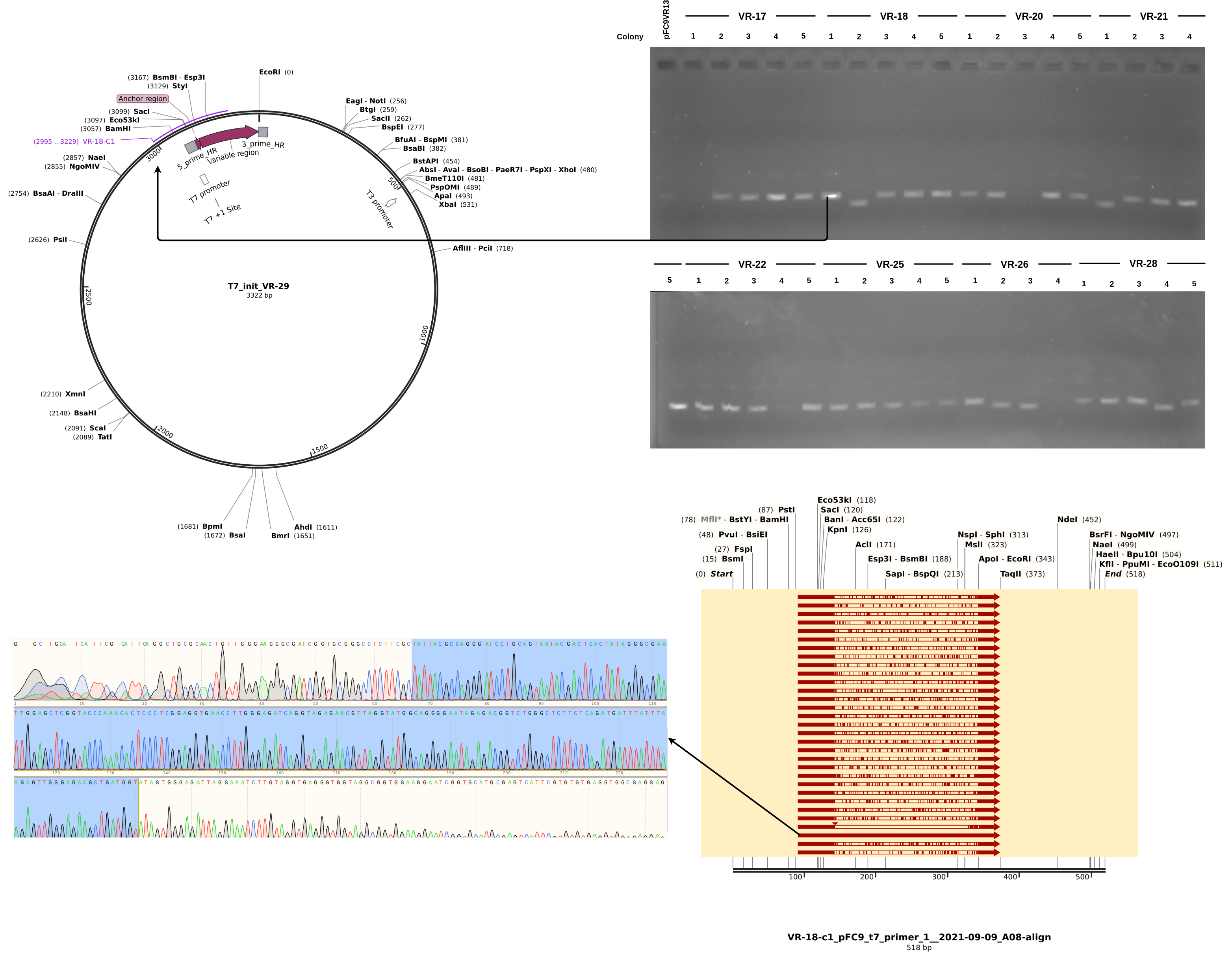
Next looking at the less successful case of VR-18-2 we see clear contamination from the DNA trace and no alignment to any VR inserts. Additionally, something a bit weird is going on with the gel in terms of the band heights. The first lane is pFC9VR13 which is confirmed to have the insert. Even though signal is faint (did not add enough DNA apparently) it is clearly at the same height as most bands except for the two that did not align to VR reference sequences; VR-28-C4 and VR-18-C2. This is indicating to me some other source of contamination in these samples but I am not sure where from.