More VR insert purifications #
Agarose purifications #
Purfied most inserts (except 29-31 due to weird PCR issues shown in next section) via agarose gel extraction using the freeze and squeeze method.
| Sample | Yield | r_260_280 | r_260_230 |
|---|---|---|---|
| VR-1 | 47.8 | 1.816 | 0.064 |
| VR-2 | 112.9 | 1.824 | 0.439 |
| VR-3 | 15.9 | 1.138 | 0.164 |
| VR-4 | 18.1 | 1.09 | 0.196 |
| VR-5 | 14.6 | 0.88 | 0.145 |
| VR-6 | 17.2 | 1.411 | 0.251 |
| VR-7 | 15.1 | 1.493 | 0.321 |
| VR-8 | 20.3 | 1.285 | 0.208 |
| VR-9 | 26.9 | 1.352 | 0.216 |
| VR-10 | 31.7 | 1.609 | 0.335 |
| VR-11 | 8.8 | 1.207 | 0.349 |
| VR-12 | 7.6 | 1.358 | 0.33 |
| VR-13 | 8.6 | 1.332 | 0.238 |
| VR-14 | 8 | 1.23 | 0.243 |
| VR-15 | 8.9 | 1.512 | 0.425 |
| VR-16 | 28.8 | 1.512 | 0.33 |
| VR-17 | 7.1 | 1.574 | 0.455 |
| VR-18 | 5.8 | 1.545 | 0.565 |
| VR-19 | 11.9 | 1.521 | 0.434 |
| VR-20 | 21.2 | 1.608 | 0.242 |
| VR-21 | 17.4 | 1.607 | 0.459 |
| VR-22 | 20.4 | 1.787 | 0.476 |
| VR-23 | 7.2 | 1.709 | 0.46 |
| VR-24 | |||
| VR-25 | 24.7 | 1.803 | 0.502 |
| VR-26 | 4.9 | 1.678 | 0.368 |
| VR-27 | 7.6 | 1.357 | 0.352 |
| VR-28 | 10.5 | 1.232 | 0.082 |
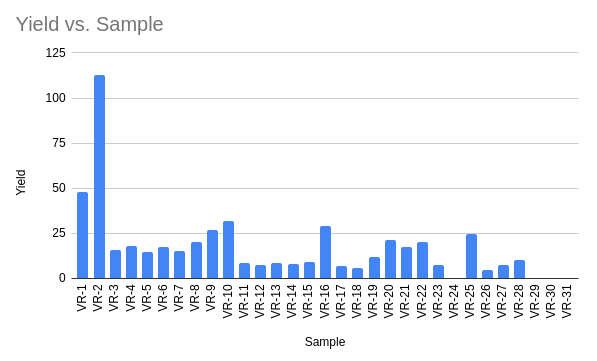
This table is also available on google drive at this link. I places all samples into the VR inserts 2 box in the kitchen freezer.
PCR VR-29, 30 and 31 #
Yesterday I did not have enough PCR master mix to amplify samples 29, 30, and 31 so I did those samples today. However, I found a Phusion polymerase master mix that was basically unused to I decided to use that instead of the OneTaq Polymerase I had used in previous reactions. Since VR-31 was supplied as a vector, after amplification I digested with BglII at 37C for 1hr. Since sample 29 and 30 were also on the same strip of tubes they were also incubated at 37C but no BglII was added to these samples. Image below is from the trans-illuminator from right to left samples are VR-29, 30 and 31.

Phusion seems to have produced more PCR product but in all cases produced a smear of DNA in addition to the main fragment band. Also it looks like BglII failed to digest the PCR product. For tomorrow will use the lab Taq polymerase and dilute the digested sample to 50ul.
Verification of Gibson assembly #
Sent Gibson assembly samples VR 3, 19, 28, and 29 for sequencing with pFC9 T7 forward primer. Then I aligned all VR sequences as ordered to Sanger data.
VR-3 #
Confirmed as pFC9VR3.

VR-13 #
Confirmed as pFC9VR13.
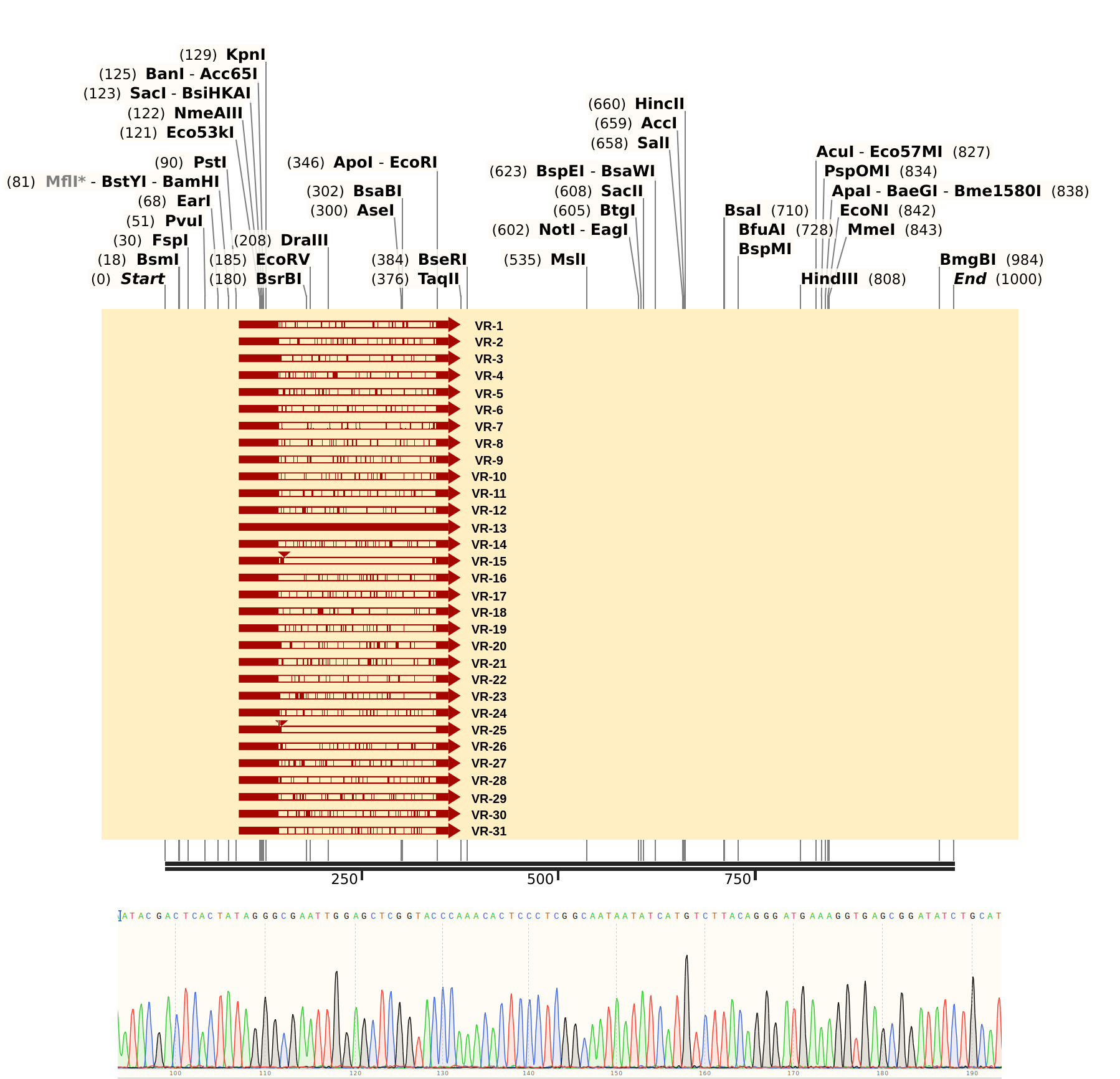
VR-28 #
Based on the Sanger trace and the lower quality of alignment it looks like sample contains pFC9VR28 but may be contaminated with something else :( will need to run gel to confirm and will likely require redoing the prep.
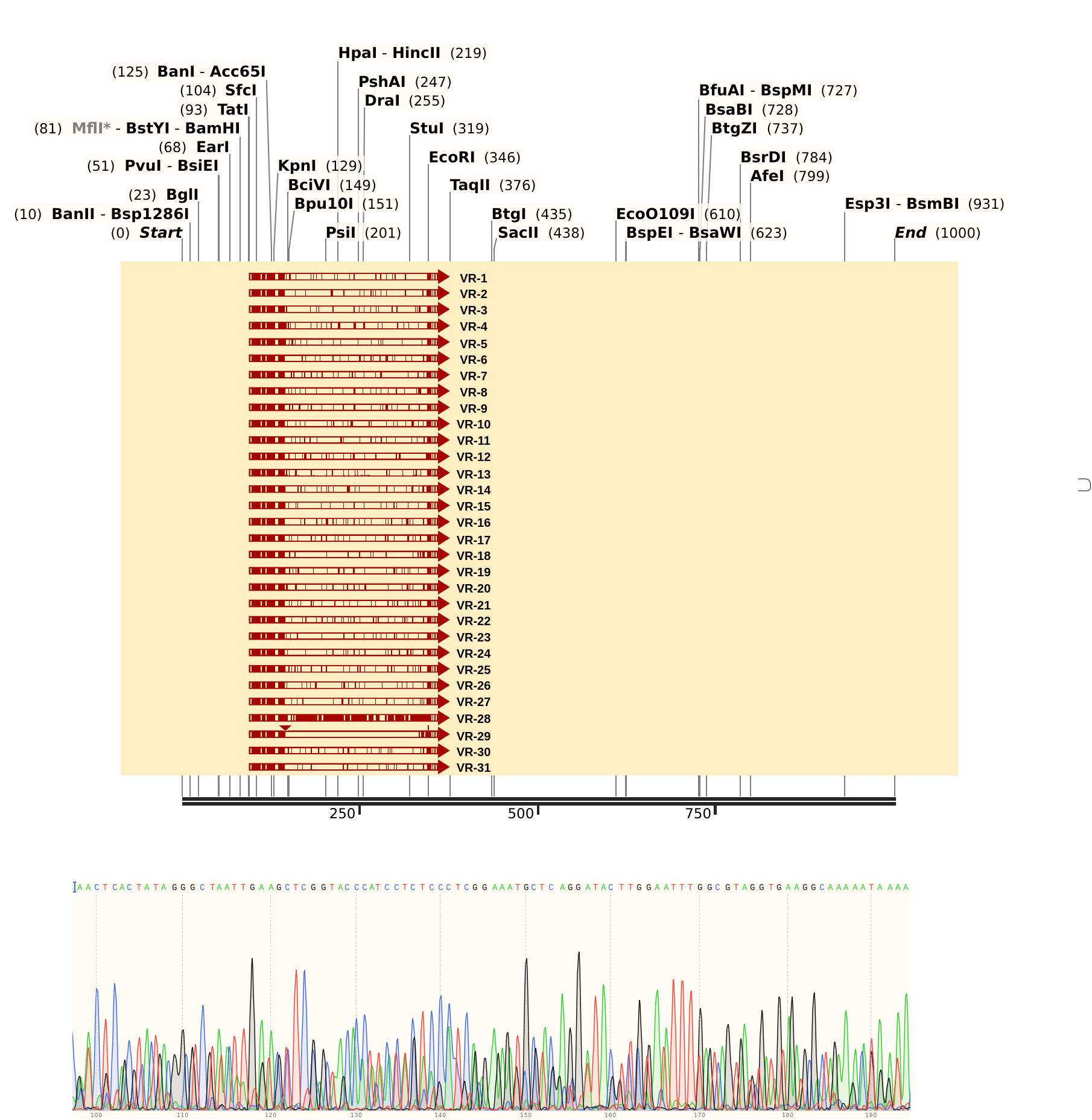
VR-29 #
Confirmed as pFC9VR29.
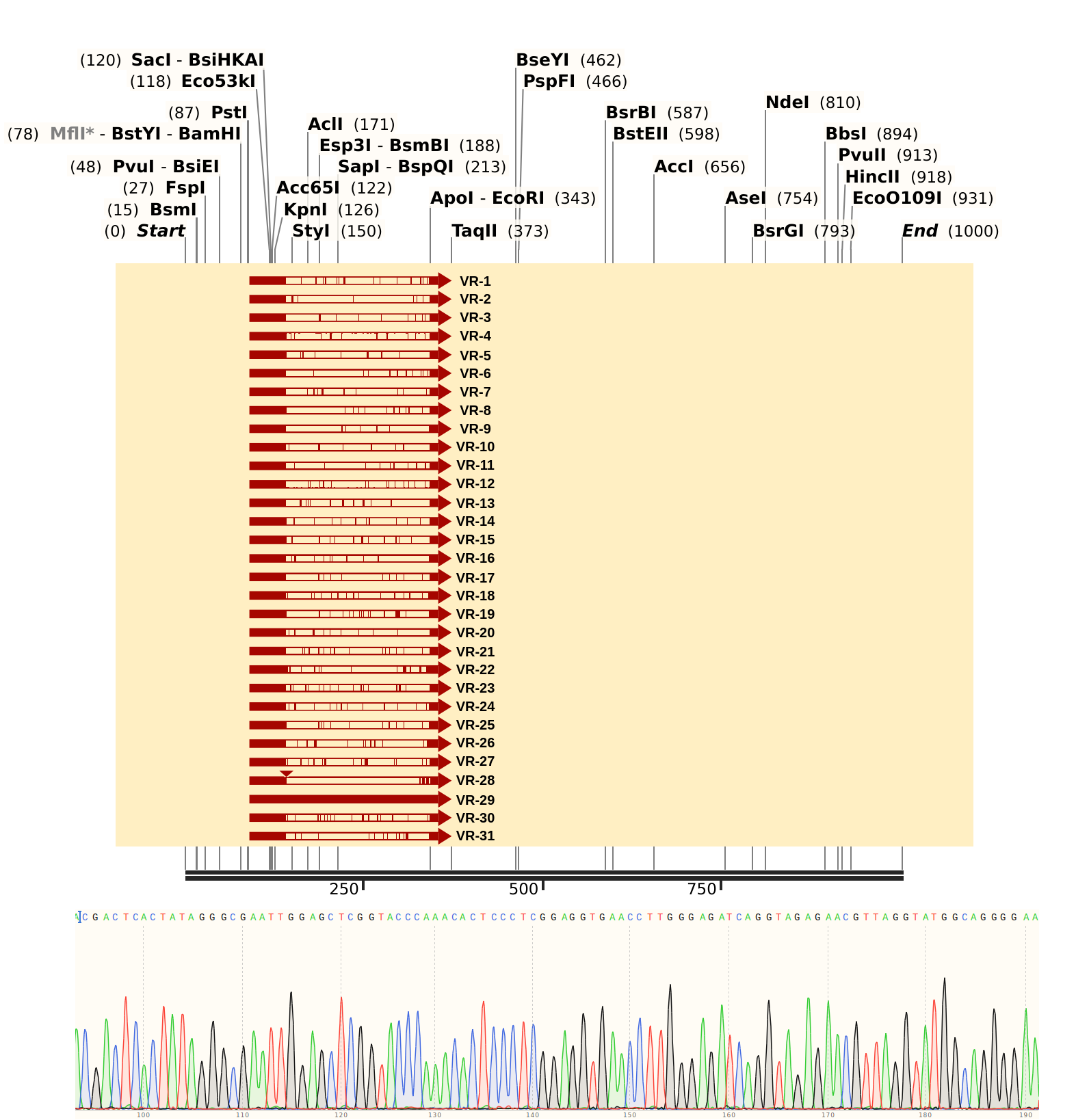
Alignment files are available in this directory.