BglII digests and one-pot PCR BglII digest #
Following up from Saturday I purchased BglII from the sci store in the morning. Today I am testing digestion of PCR vector fragments with BglII and accessing if it is possible to do a BglII digestion followed by PCR in the same tube.
BglII digests #
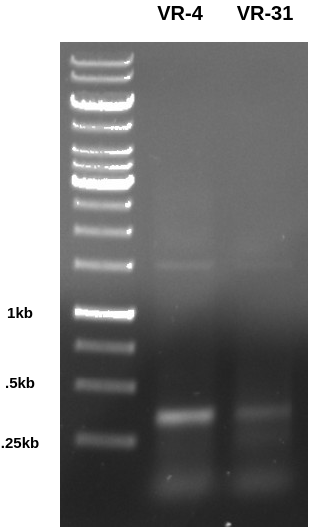
First set up a PCR reaction with VR-4 and VR-31 to create amplicons that contain the repeated homology motif and subsequently one BglII site. Ran the gel below (0.8 agarose in TAE at 120V for 45 mins) to confirm that the PCR worked correctly.
After confirming PCR worked I digested alliquotes of each sample with BglII overnight at room temperature (was leaving the lab at the time reaction was ready to go). I then ran the gel the next day (I know this is slightly out of order but makes more sense to put in these notes.) The gel below was run at 150V because I was in a hurry to get out of the lab (was getting late) and this ended up causing a pretty annoying amount of smearing. In the future I will max out voltage at 130 with 120 being a normal running voltage and 100 being for extra special gels.
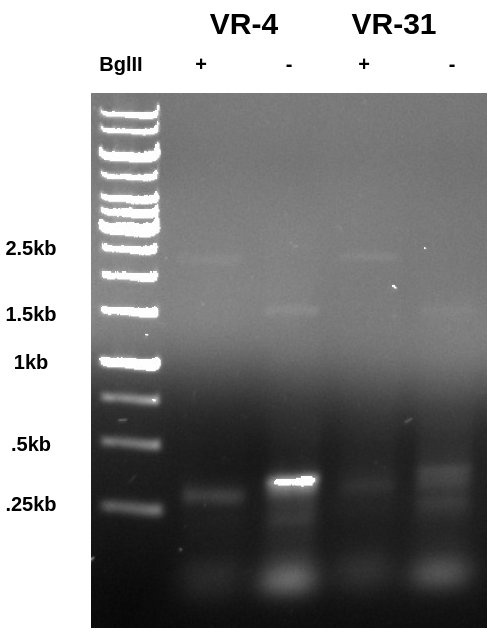
In the BglII digested lane the PCR product is shifted downwards indicating the BglII was capable of digesting the PCR product despite the close proximity of the recognition sequence to the end of the fragment. Additionally if you look closely you can see a band at 1.5kb in the undigested lane that seems to shift up to 2.5kb in the digested lane. This is the vector which is intact and supercoiled in the undigested samples but is then linearized in the BglII digested lane.
pFC9 SacI EcoRI large fragment extraction “Freeze and squeeze” #
I wanted to test the freeze and squeeze method of DNA extraction from agarose gels. To test I selected two unpurified pFC9 SacI EcoRI digestions from 8/17/21
Extraction results #
Yield was modest but considering the overall volume acceptable for a first attempt. I think some amount of DNA was lost when loading the gel, as I was really trying to squeeze sample into small wells and I noticed at least some degree of overflow / spilling when loading the wells and moving the box over to the power supply.
| Sample | ng/ul |
|---|---|
| 1-1 | 20.6 |
| 1-2 | 6.9 |
| 2-1 | 3.7 |
| 2-2 | 3.2 |
I then ran 10 ul aliquots of each sample out on a gel.
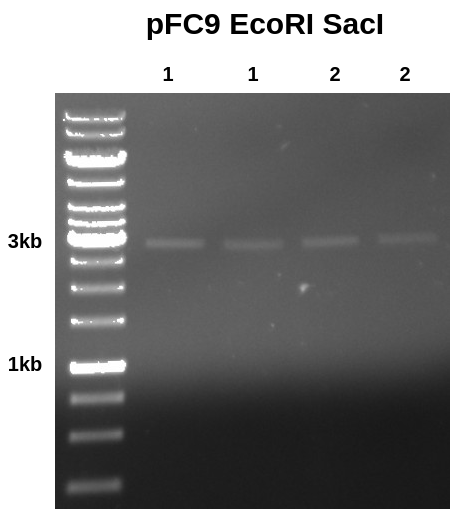
Results looks clean with band where it should be, additionally the intensity of each band does not look that different to me despite the nanodrop measuring sample 1-1 (lane 1) to have about 4x the amount of DNA as all other samples.
PCR BglII digest experiments #
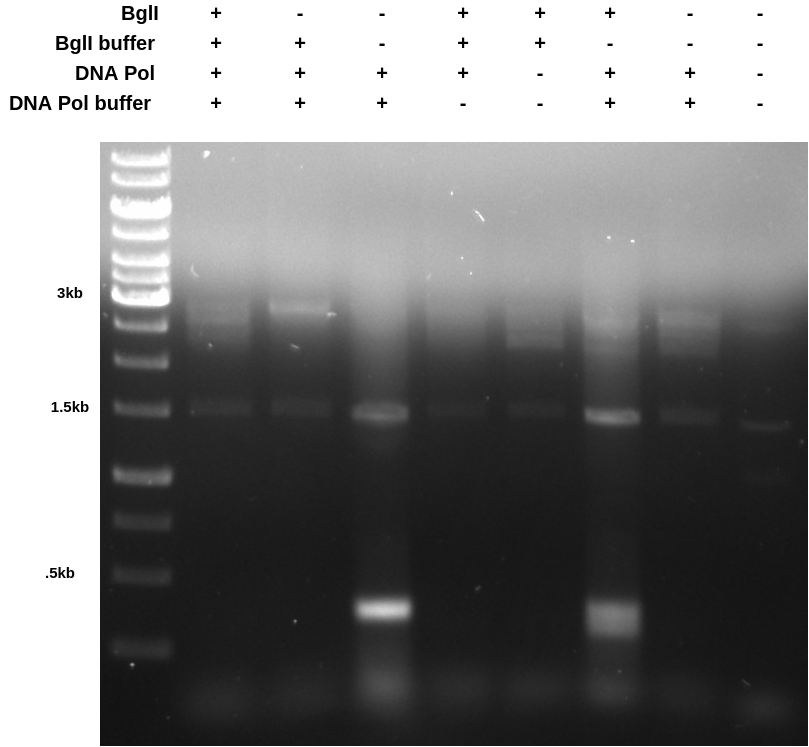
These results were somewhat confusing but it seems to me that DNA polymerase may not be compatible with the BglII buffer. In the future I will just run the PCR reaction first, then dilute the reaction to a total volume of 50 ul and and BglII and buffer.