VR-pFC9 Gibson assembly and Sanger seq analysis #
Today I am using the VR PCR fragments I generated on 8/18/21 and pFC9 large fragment sample 1 generated on 8/17/21 to attempt to create pFC9 T7 initiation series constructs.
Gibson assembly reagents #
I created a 5x isothermal buffer and 1.33x Gibson assembly master mix according to the values described in this spreadsheet. This spreadsheet is also where I will be stored all future Gibson assembly reactions.
VR inserts #
Inserts used for each reaction are listed in the table below.
| Fragment | Length | Concentration (ng/ul) | Sample number |
|---|---|---|---|
| VR-7 | 281 | 151.6 | 1 |
| VR-13 | 281 | 108 | 2 |
| VR-3 | 281 | 166.6 | 3 |
| VR-5 | 281 | 120.5 | 4 |
| VR-18 | 281 | 81.7 | 5 |
| VR-29 | 281 | 149.9 | 6 |
| VR-10 | 281 | 199.9 | 7 |
| VR-28 | 281 | 39.2 | 8 |
Gibson assembly reactions #
All reagents for each reaction are described in this spreadsheet. All samples
were incubated at 50C for 1hr. Extra Gibson reaction volumes were stored in the VR inserts box. Image of
all samples below.
Transformation in chemically competent E. coli #
Picked up chemically competent E. coli per Fred’s recommendation from Davis Scientific store. They only had one box of expired cells which I took but was not charged for (nice). Followed transformation protocol that was included with the cells and utilized the provided positive control DNA in addition to my own samples. Each aliquot of cells was transformed with 5ul of Gibson reaction.

Plated each sample onto agar with Amp and placed into 37C incubation room at ~4:30 pm.

Additional transformed cells were placed at 4C in the deli fridge for possible use tomorrow as per the included chemically competent cell protocol.

VR fragment and vector sanger results #
On Friday I submitted a number of samples for Sanger sequencing to double check Thermo’s work and ensure correct insert sequence was included in each vector.
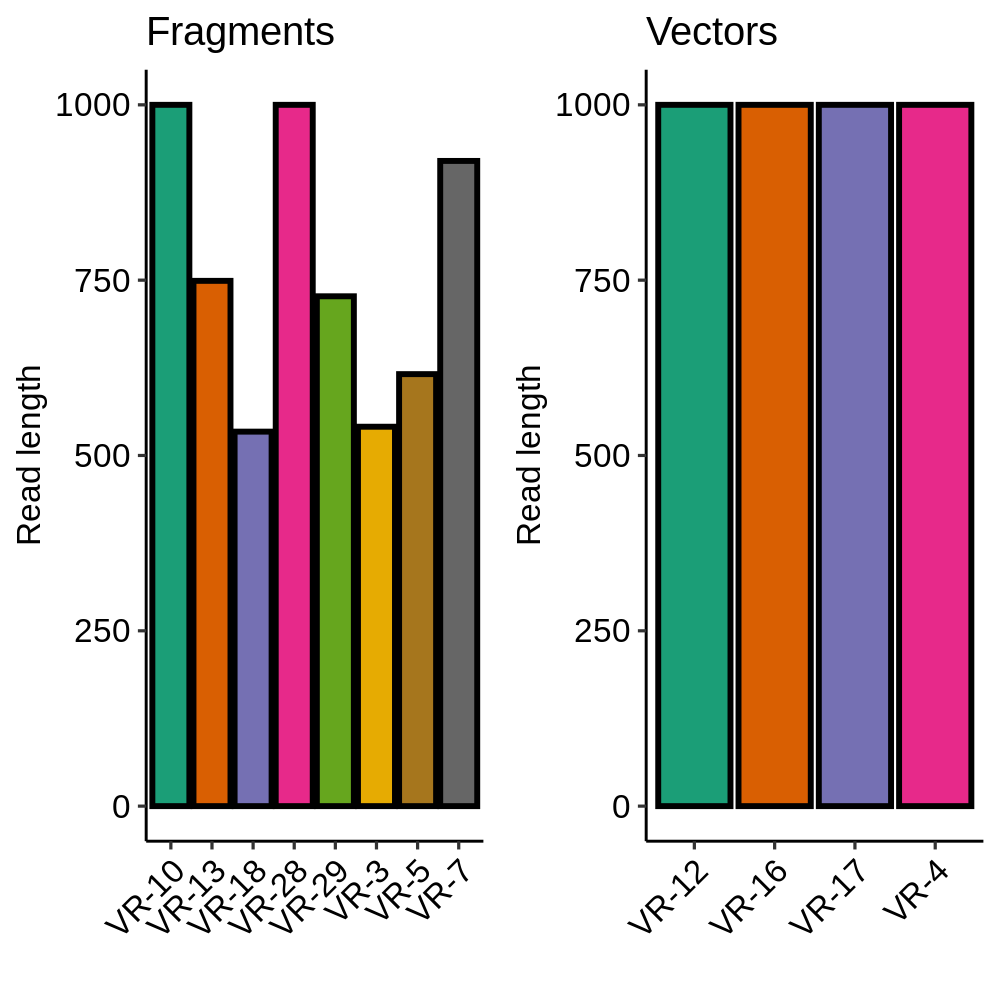
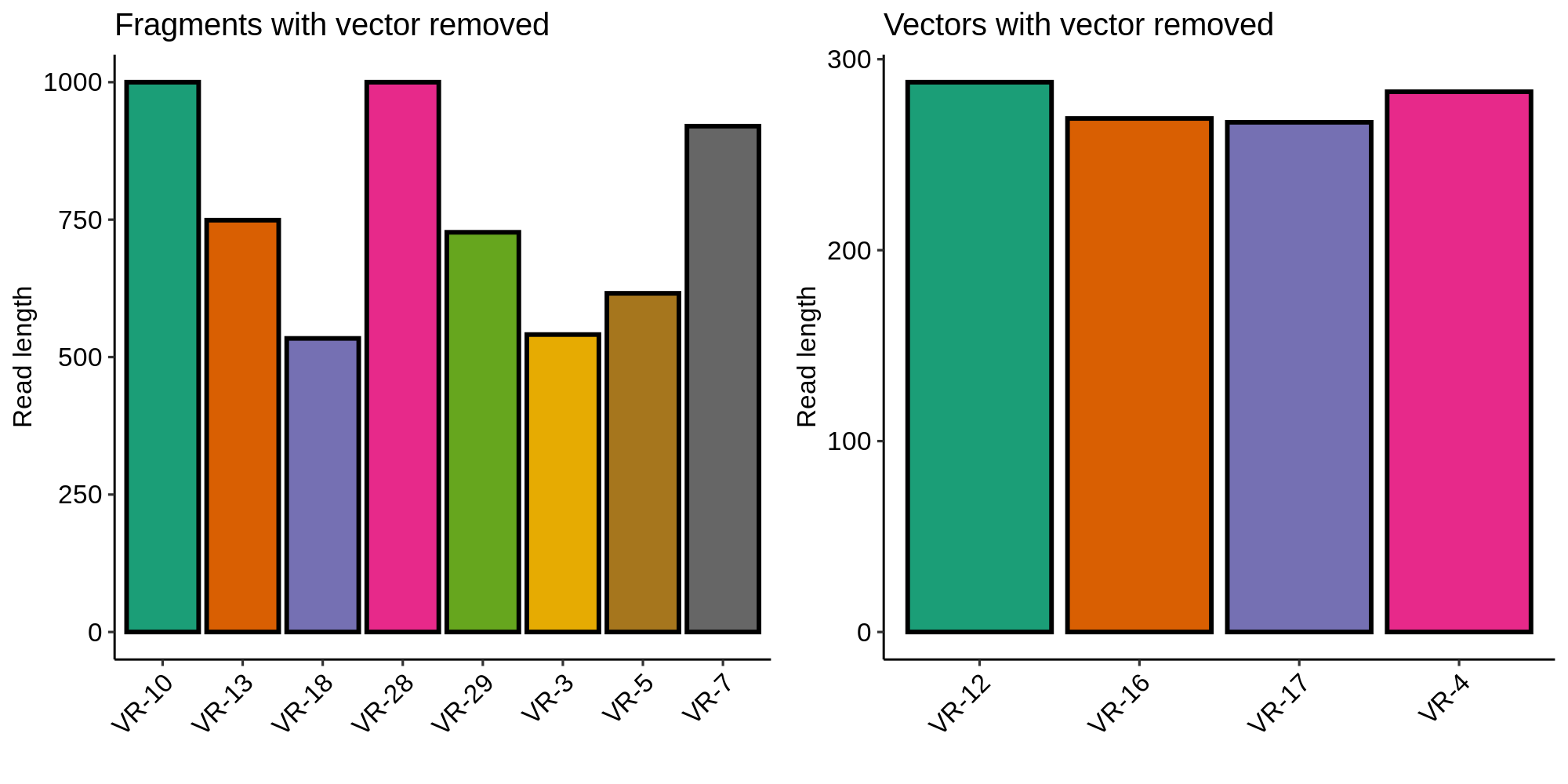
Removing vector sequences corrected the lengths of inserts included as part of a vector and did not affect fragments (as expected). This also ruled out contamination of the long fragments with vector sequence.
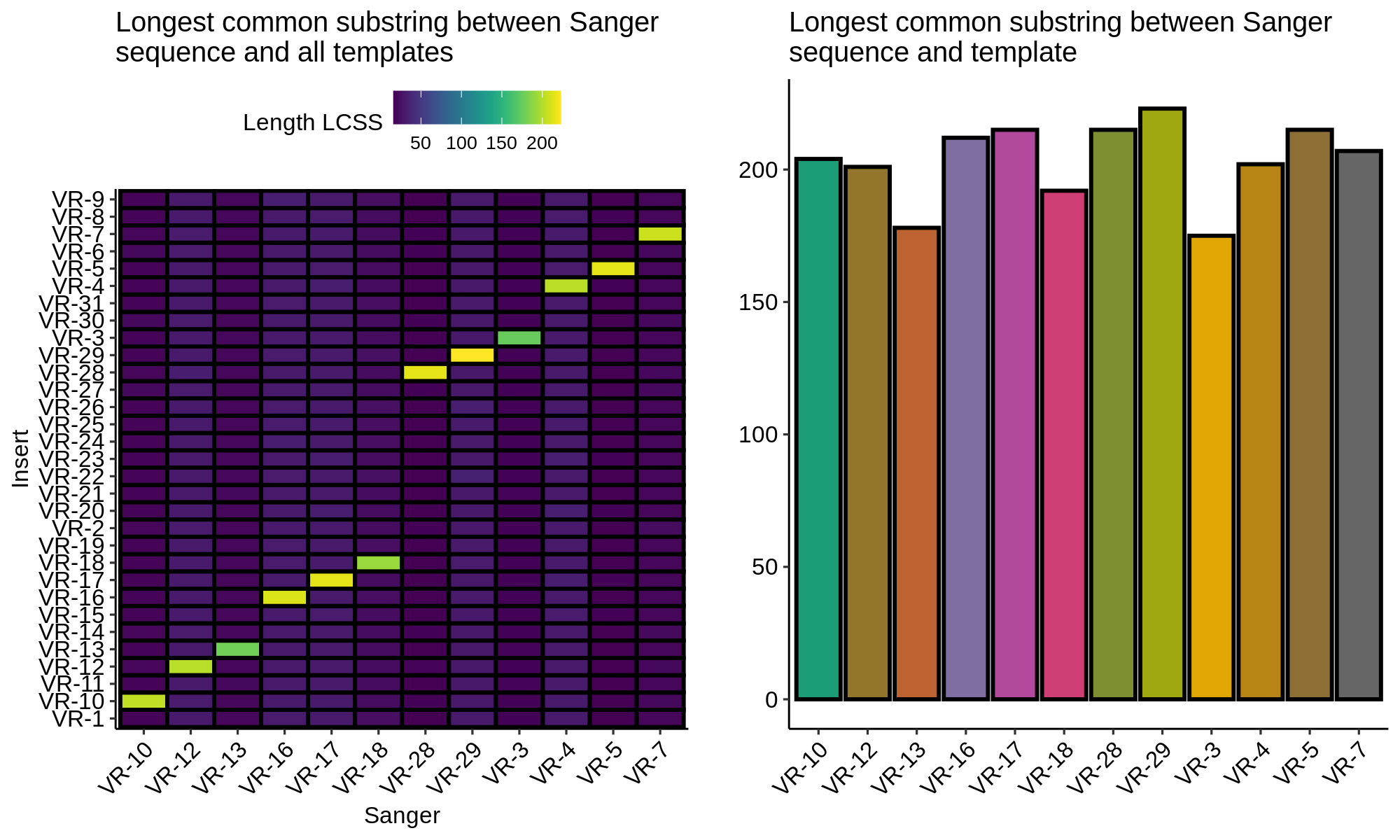
Finding the longest common substring between Sanger sequences and VR insert templates showed that all sequences are labeled correctly and contain the correct sequences.
However this still left the question of why the fragments were more than double the length than I was expected them to be based on the gel I ran before submitting them for sequencing. What I should have done first was check the entire chromatogram for at least a couple of the sequences.
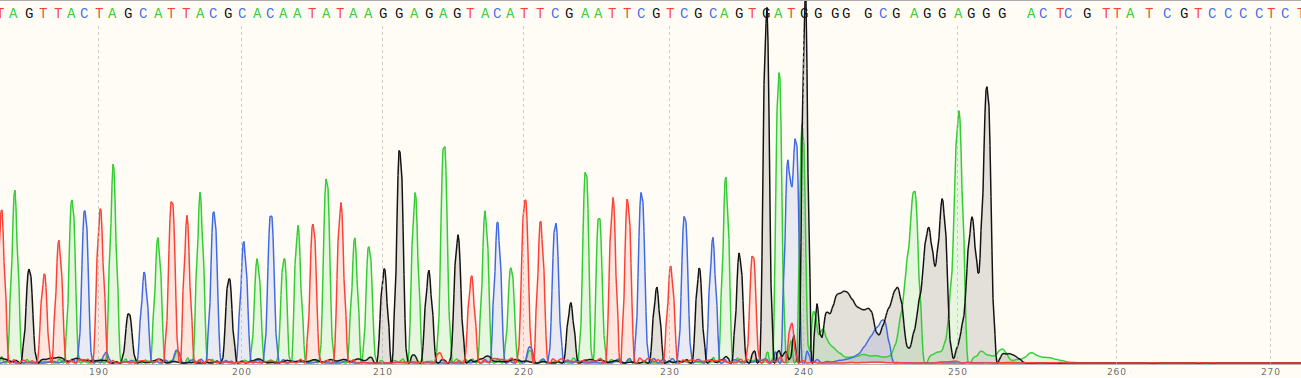
As you can see in the image above from VR-10 the signal completly dies after ~260 bp. Which is just about the length we would expect given the primer used. Overall the conclusion is that the gel showing different lengths for VR-7 and VR-8 was weird, but sequencing shows that everything is as expected. The Jupyter notebook and the seqeuncing data used to generate the plots above can be found in notebooks/VR-insert-sanger-verification. Sanger sequencing data is also available in Google drive at this link.