VR inserts quality checks continued #
PCR results #
Ran PCR from yesterday out on 0.8 agarose TAE gel at 120V for 1hr.
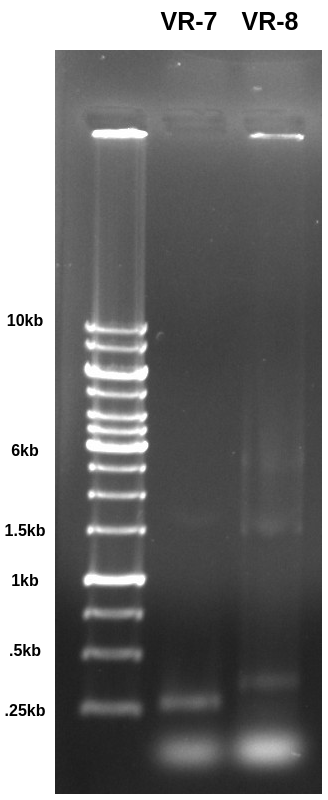
VR-7 is a fragment while VR-8 is a included as part of a vector. Both were PCR amplified with primers that should produce ~280bp fragments. The band is basically right where I would expect it for the VR-7 sample but a but higher for VR-8 which is making me think maybe something else is included when in the vector form? Not sure about that. Need to digest with BglII and see what the results look like.
Also it seems like a lot of something got stuck in both the ladder and VR-8 lanes. Not sure exactly what that signal is coming from and why the it would not be seen in the VR-7 lane.
PCR of VR$_n$ fragments for sequencing #
Given the results in the gel above, I am planning on submitting several VR fragments and vectors for Sanger sequencing to “trust but verify” and hopefully determine what may be causing the discrepancy in lengths. While there is plenty of vector inserts to send directly for sequencing, there is far less fragment to go around. So I am using the VR insert targeting primers to amplify 8 fragments and then will clean up the results and send the amplicons for sequencing.
PCR reactions #
PCR reactions and reagents are recorded in this spreadsheet. VR inserts that were amplified and their sample numbers are listed in the table below.
| DNA template | Concentration (ng/ul) | ng DNA per reaction | Sample number |
|---|---|---|---|
| VR-7 | 50 | 15 | 1 |
| VR-13 | 50 | 15 | 2 |
| VR-3 | 50 | 15 | 3 |
| VR-6 | 50 | 15 | 4 |
| VR-18 | 50 | 15 | 5 |
| VR-29 | 50 | 15 | 6 |
| VR-10 | 50 | 15 | 7 |
| VR-28 | 50 | 15 | 8 |
After PCR was completed, cleaned up each reaction by following the Zymogen DNA cleanup kit protocol. Then I nanodroped all samples results are shown in the table below
| Insert | ng/ul | 260/280 | 260/230 | Sample volume (ul) |
|---|---|---|---|---|
| VR-7 | 151.6 | 2.195 | 2.857 | 25 |
| VR-13 | 108 | 2.139 | 3.487 | 25 |
| VR-3 | 166.6 | 2.33 | 2.68 | 25 |
| VR-6 | 120.5 | 2.171 | 2.802 | 25 |
| VR-18 | 81.7 | 2.033 | 2.112 | 25 |
| VR-29 | 149.9 | 2.175 | 2.401 | 25 |
| VR-10 | 199.9 | 2.338 | 2.721 | 25 |
| VR-28 | 39.2 | 1.989 | 2.418 | 25 |
Overall I am very happy with these results. There is plenty of DNA to prepare for sequencing tomorrow.
I put all samples into the VR inserts box on my shelf in the kitchen freezer. Image of the samples is shown below.

Vector inserts #
While I did not do any prep for these because they have much higher concentrations then fragment inserts I will prep VR 12, 17, 27, and 4 for sequencing tomorrow.
Gibson assembly reagent prep #
For preparing insert vectors I think it would be best to make my own Gibson reagents as buying the premixed stuff comes out to a pretty significant cost per reaction. I created this Gibson assembly spreadsheet to calculate reagent amounts for both isothermal buffer and the master mix. Typical values for 5x isothermal buffer and Gibson master mix are included in the tables below.
5x Isothermal buffer.
| Reagent | Concentration | Unit | Volume stock (ul) |
|---|---|---|---|
| Tris-HCL | 500 | mM | 625 |
| MgCl2 | 50 | mM | 125 |
| PEG-8000 | 25 | % | 833.3333333 |
| NAD | 5 | mM | 250 |
| dNTPs | 1 | mM | 250 |
| PCR grade H20 | NA | NA | 416.6666667 |
| Grand total (ml) | 2.5 |
Gibson assembly master mix.
| Reagent | Concentration | Unit | Volume stock (ul) |
|---|---|---|---|
| Taq ligase | 5 | units/ul | 46.875 |
| 5x isothermal buffer | 1 | X | 75 |
| DNA polymerase | 0.2 | units/ul | 15 |
| T5 exonuclease | 0.005 | units/ul | 0.1875 |
| PCR grade H20 | NA | NA | 237.9375 |
| Grand Total | 375 | ||
| Grand total (ml) | 2.5 |
Sanger sequencing results and pFC identification #
Got sequencing results that were submitted yesterday and prepared on Tuesday back. Raw data is stored in google drive and is available at this link.
A summary of the results is below.
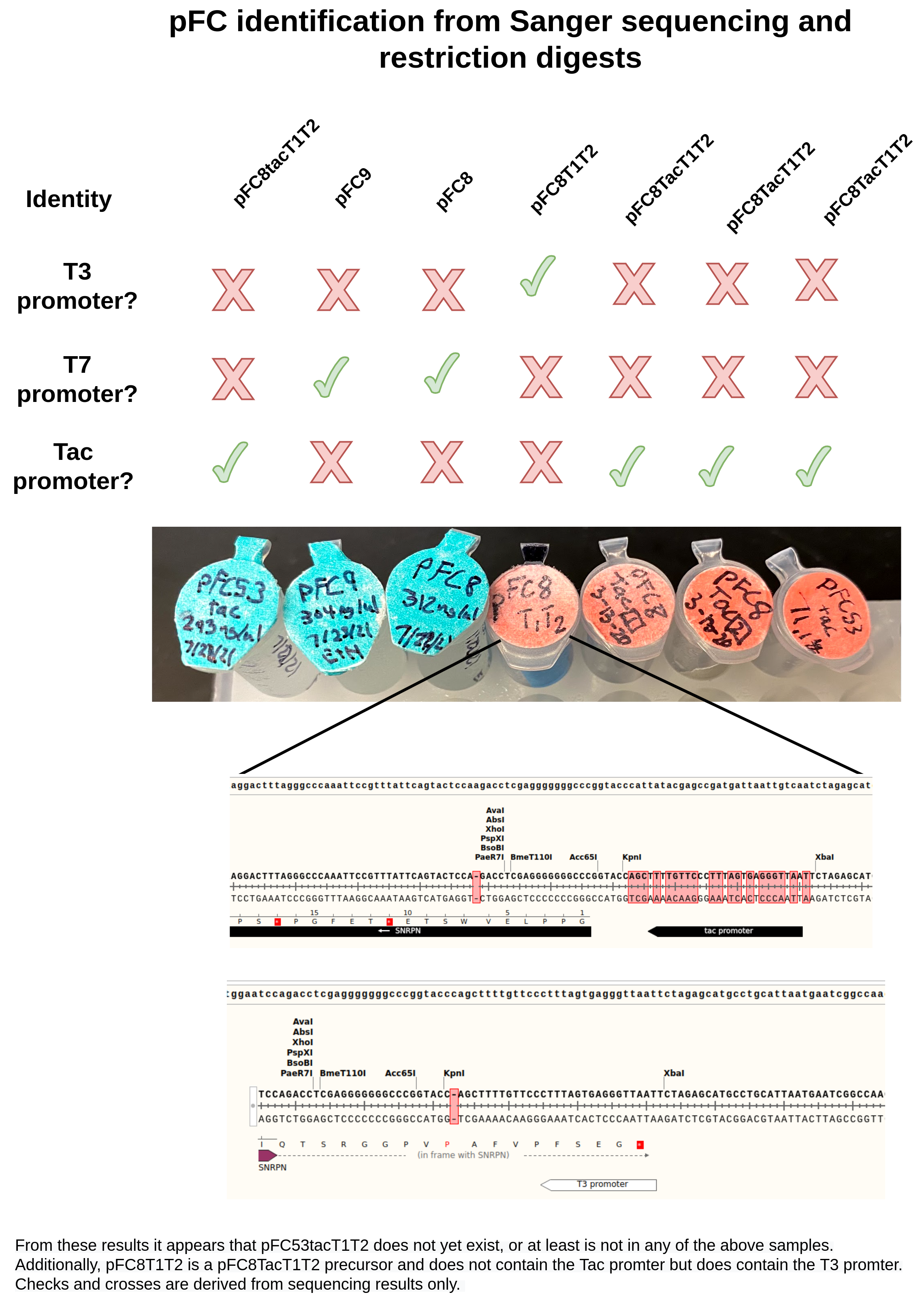
Alignment of pFC8 and pFC9 samples to pFC9 confirmed their respective identities. In image below pFC8 is the blue arrow while pFC9 sample is the red arrow.