pFC9 digest with EcoRI SacI #
Back from vacation and the inserts are on their way to Davis. Today I am preparing more pFC9 large fragment for cloning via digestion with SacI and EcoRI and agarose gel extraction.
pFC9 digest protocol #
Digest 1 ul pFC9 for 1.5 hours with SacI, then digest with EcoRI for 0.5 hrs. Incubate at 37C in PCR machine. Extract large fragment using Zymoclean DNA recovery protocol.
Reagents #
Split into 5 separate reactions for incubation. Total DNA per reaction is then 1 ug.
| Reagent | Volume (ul) |
|---|---|
| pFC89 | 16.4 |
| SacI-HF | 5 |
| EcoRI-HF | 5 |
| Cutsmart buffer | 25 |
| H20 | 198.6 |
Agarose gel pours #
Poured agarose gel very thin, 25 ul per gel. Hopefully will reduce amount of gel during extraction but probably need to split samples as I don’t think the 50ul volume will fit into one lane.
Tested gels after set with loading dye to measure capacity and was only about 15 ul per lane. Re-poured gels with similar depth but larger wells (6 per gel).
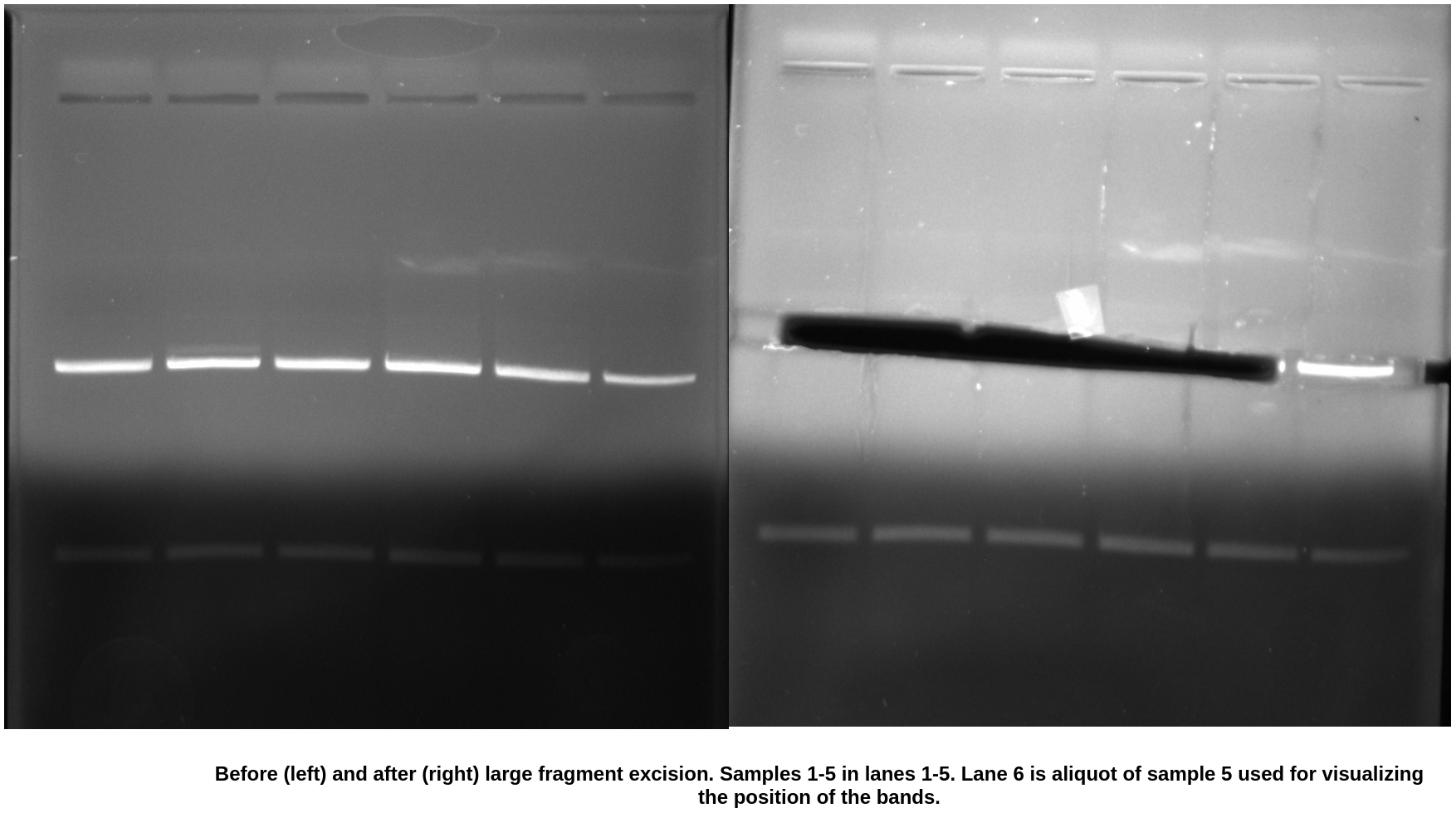
Gel extraction #
Gel extraction went pretty well. Bit heavier on the agarose then I would have liked but this is the cost of using the large wells even with thin pours.
| Sample | Agarose mass (g) |
|---|---|
| 1 | 0.154 |
| 2 | 0.173 |
| 3 | 0.165 |
| 4 | 0.150 |
| 5 | 0.147 |
Nanodrop results #
| Sample | DNA ng/ul | 260/280 | 260/230 | Volume (ul) | DNA mass (ng) |
|---|---|---|---|---|---|
| 1 | 27.1 | 1.226 | 0.027 | 14 | 379.4 |
| 2 | 29.3 | 1.285 | 0.027 | 14 | 410.2 |
| 3 | 21.7 | 1.116 | 0.040 | 14 | 303.8 |
| 4 | 8.6 | 1.333 | 0.043 | 14 | 120.4 |
| 5 | 10.5 | 1.117 | 0.063 | 14 | 147 |
While the ratios are still similar to previous results the efficiency of recovery is definitely better. I am pretty happy with these results and plan to use these samples for cloning moving forward.
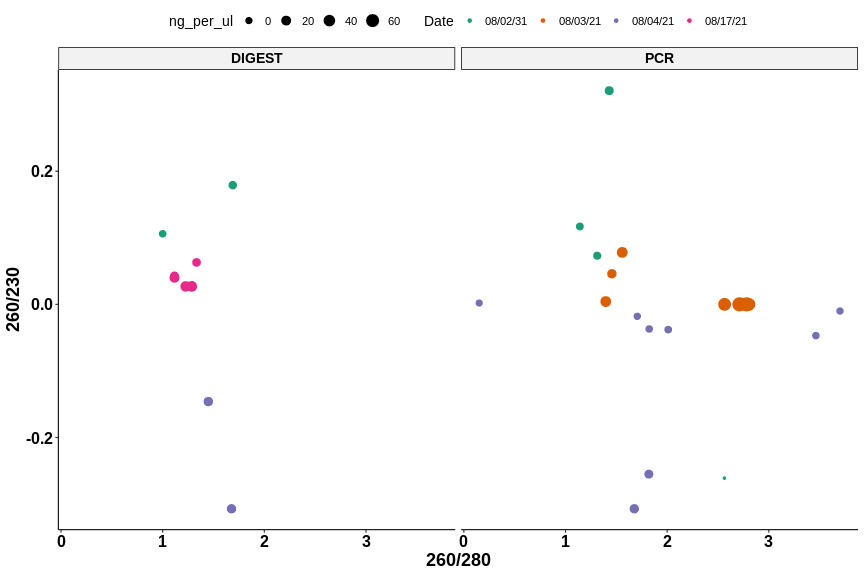
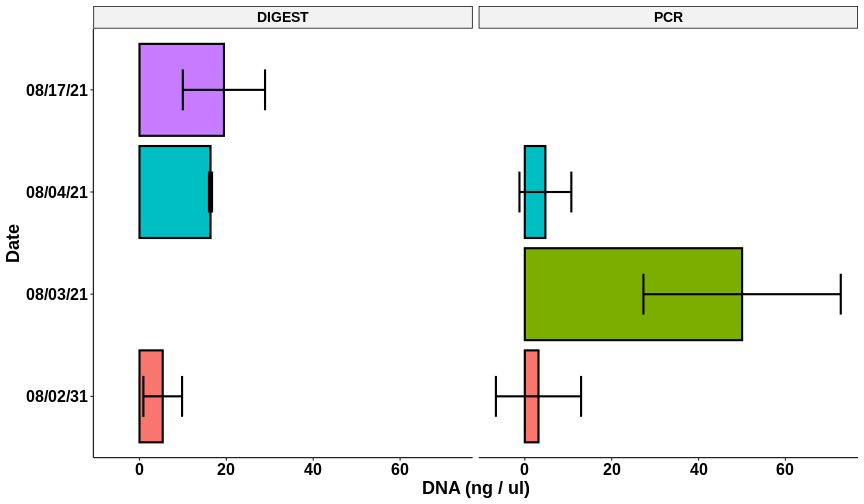
Plots produced using this Jupyter notebook.
Image of all completed samples. All samples placed into the VR-inserts box.
Sanger prep #
I also prepared a number of pFC$_n$ samples for Sanger sequencing. This was the first time I have done this so I have complied a list of notes for future sequencing jobs.
- The lab generally used Quintara Bio for all Sanger work. They have a pickup box in LSB Monday, Wednesday and Friday. Pick up time is 5pm.
- When prepping samples only include one primer.
I created this google sheet for Sanger sequencing sample prep calculations and includes the samples I prepped today but this information is also included below.
Sanger samples #
Following samples were included for sequencing.
| Plasmid | Concentration (ng / ul) | Forward primer | Forward primer concentration |
|---|---|---|---|
| pFC53tac(EH) | 300 | pFC8tac_tac_promoter_Primer_1 | 10 |
| pFC9(EH) | 300 | pFC9_t7_primer_1 | 10 |
| pFC8(EH) | 293 | pFC9_t7_primer_1 | 10 |
| pFC8T1T2(RL) | 250 | pFC8tac_tac_promoter_Primer_1 | 10 |
| pFC8tacI(RL) | 700 | pFC8tac_tac_promoter_Primer_1 | 10 |
| pFC8tacII(RL) | 320 | pFC8tac_tac_promoter_Primer_1 | 10 |
| pFC53T1T2(RL) | 123 | pFC8tac_tac_promoter_Primer_1 | 10 |
| Plasmid | Plasmid volume (ul) | H20 | Forward primer volume (ul) | Total volume | Label |
|---|---|---|---|---|---|
| pFC53tac(EH) | 4 | 8.5 | 2.5 | 15 | 1 |
| pFC9(EH) | 4 | 8.5 | 2.5 | 15 | 2 |
| pFC8(EH) | 4.09556314 | 8.40443686 | 2.5 | 15 | 3 |
| pFC8T1T2(RL) | 4.8 | 7.7 | 2.5 | 15 | 4 |
| pFC8tacI(RL) | 1.714285714 | 10.78571429 | 2.5 | 15 | 5 |
| pFC8tacII(RL) | 3.75 | 8.75 | 2.5 | 15 | 6 |
| pFC53T1T2(RL) | 9.756097561 | 2.743902439 | 2.5 | 15 | 7 |
Image of all samples from right to left same order as table above.
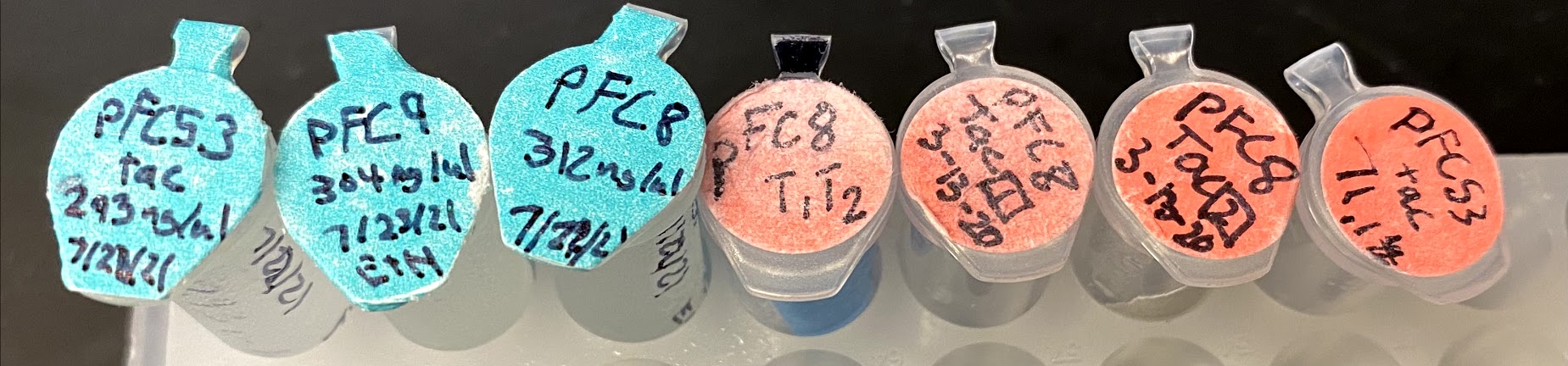
Order information #
Samples are only picked up on Monday, Wednesday and Friday so I placed into freezer for storage overnight. The order form included with the samples is at this link.