Sanger results analysis #
BLAST alignment heatmap #
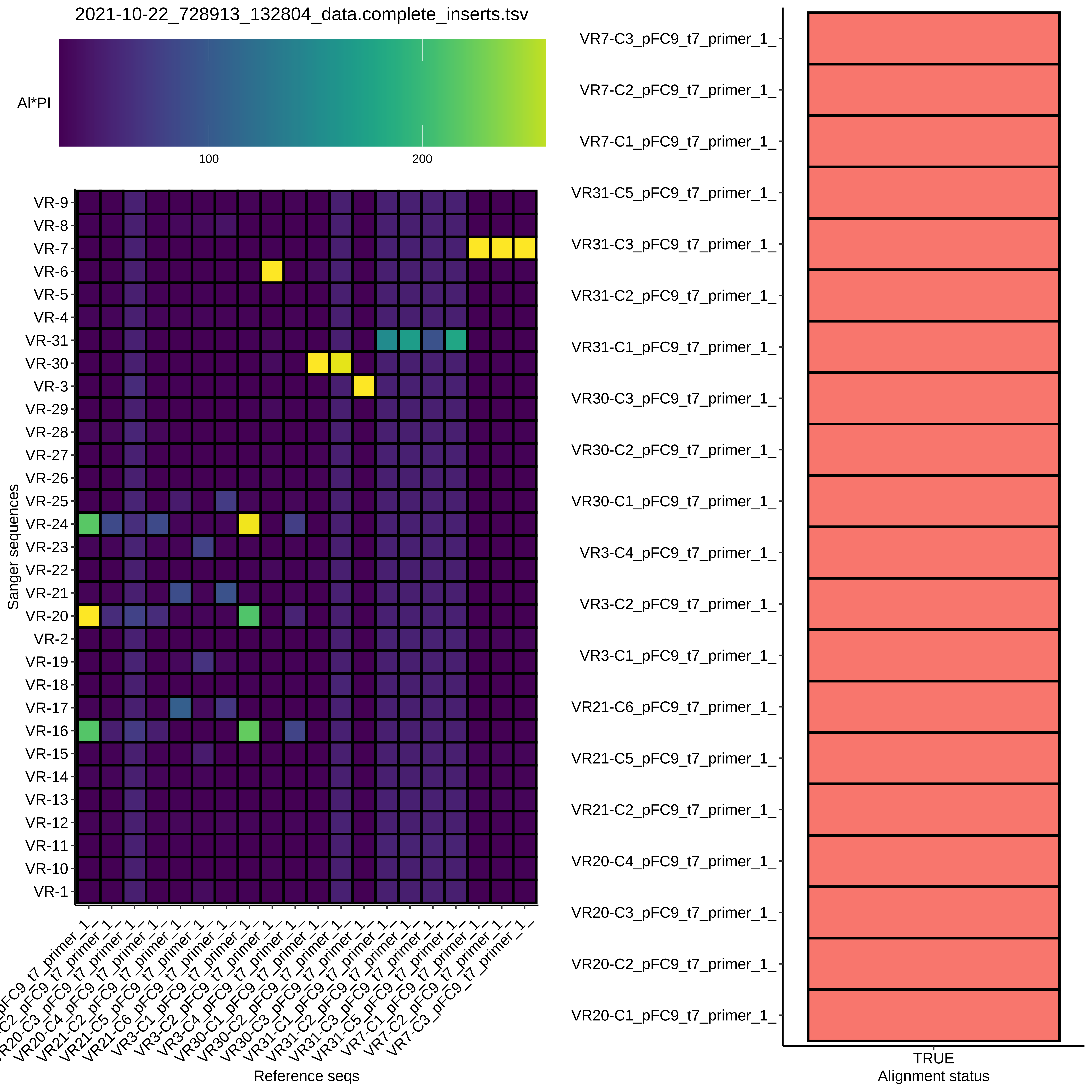
Not quite as clean as I was hoping for, but some promising results.
VR3 #
From heatmap alignment alignments of VR-3 sample to VR-3 references seem to be weak.
C1 #
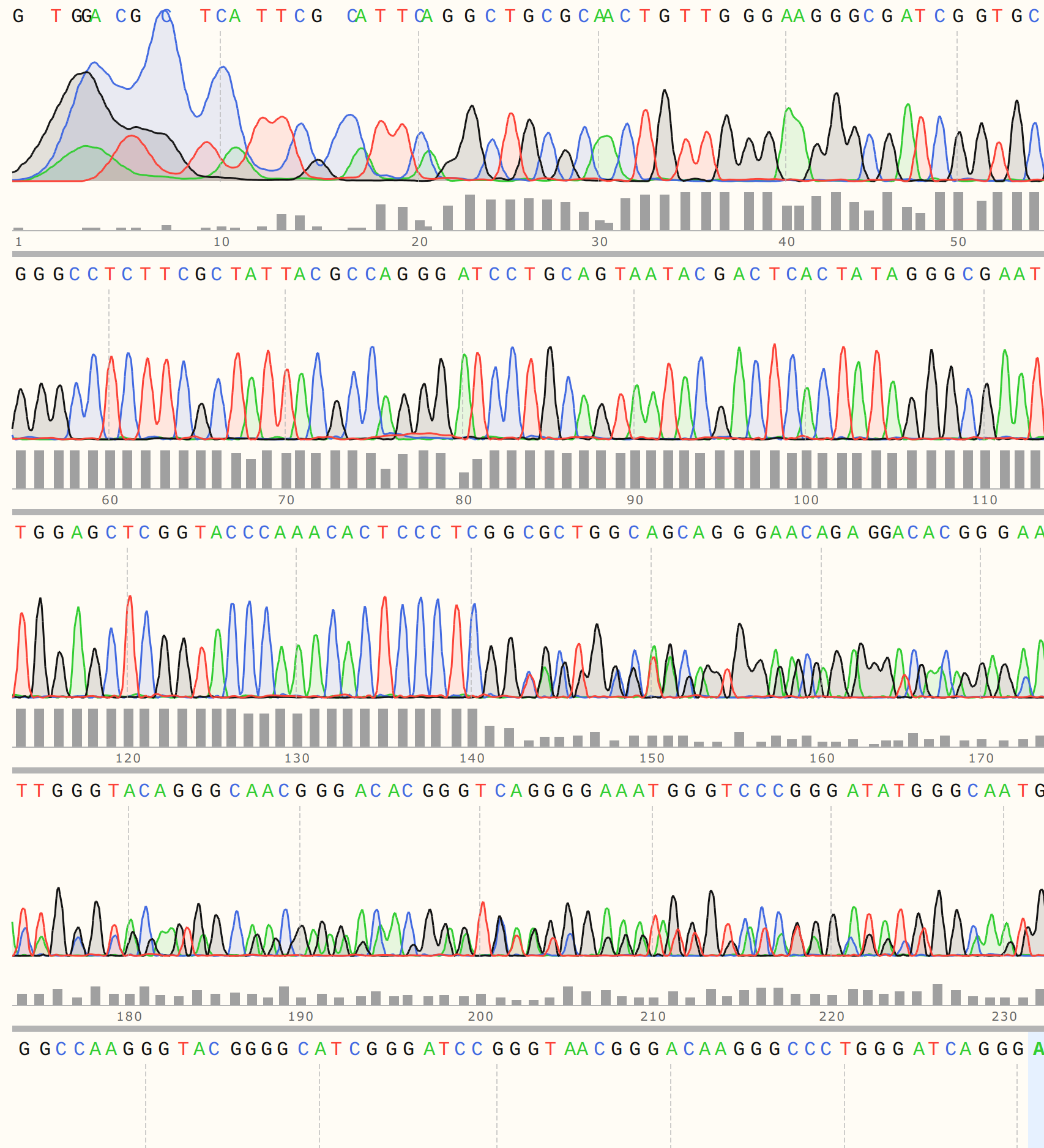
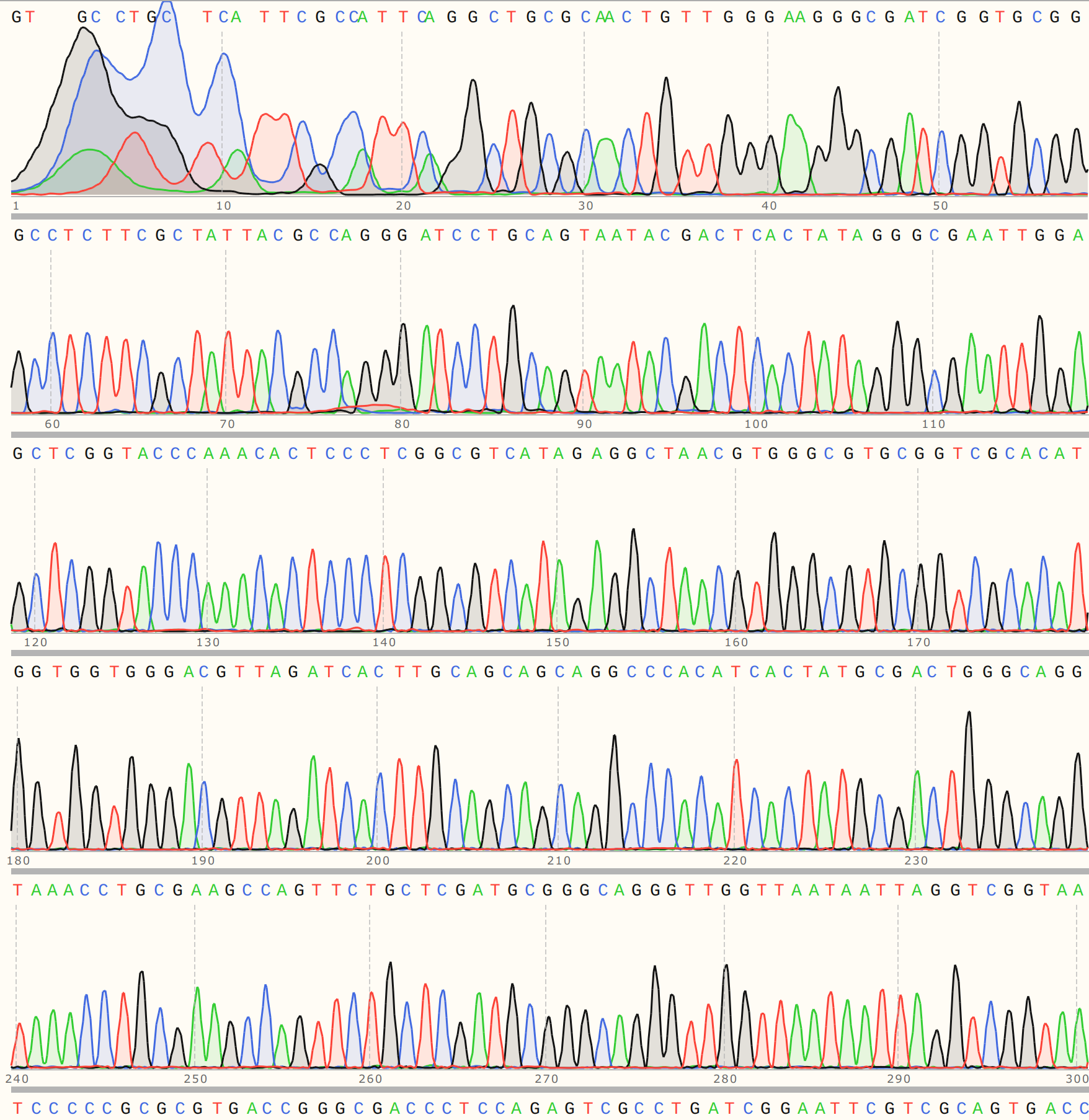
Looking at the trace shows that there appears to be relatively strong signal until around 140 bp out where things start to break down. This is just about the distance from the primer binding site to where the variable region of the VR insert should start.
There is likely a mix of more than one insert in this sample. Should proceed by “purification by cloning”.
C2 #
Colony 2 shows shows strong alignment to VR-6 in the heatmap. This holds when checking the detailed alignment via snapgene.
 . The trace
also looks strong for this sample, especially compared to C1.
. The trace
also looks strong for this sample, especially compared to C1.
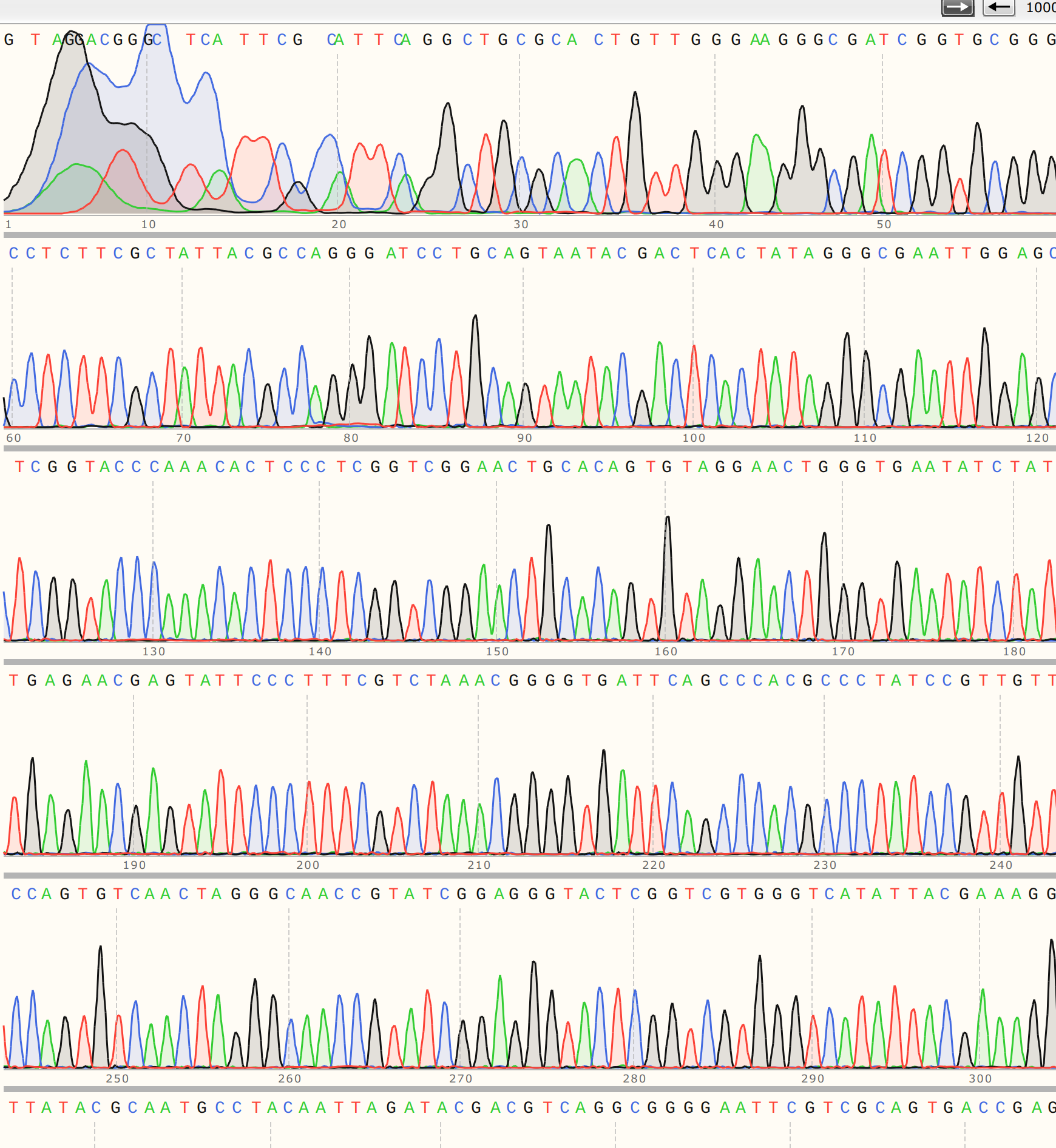
C4 #
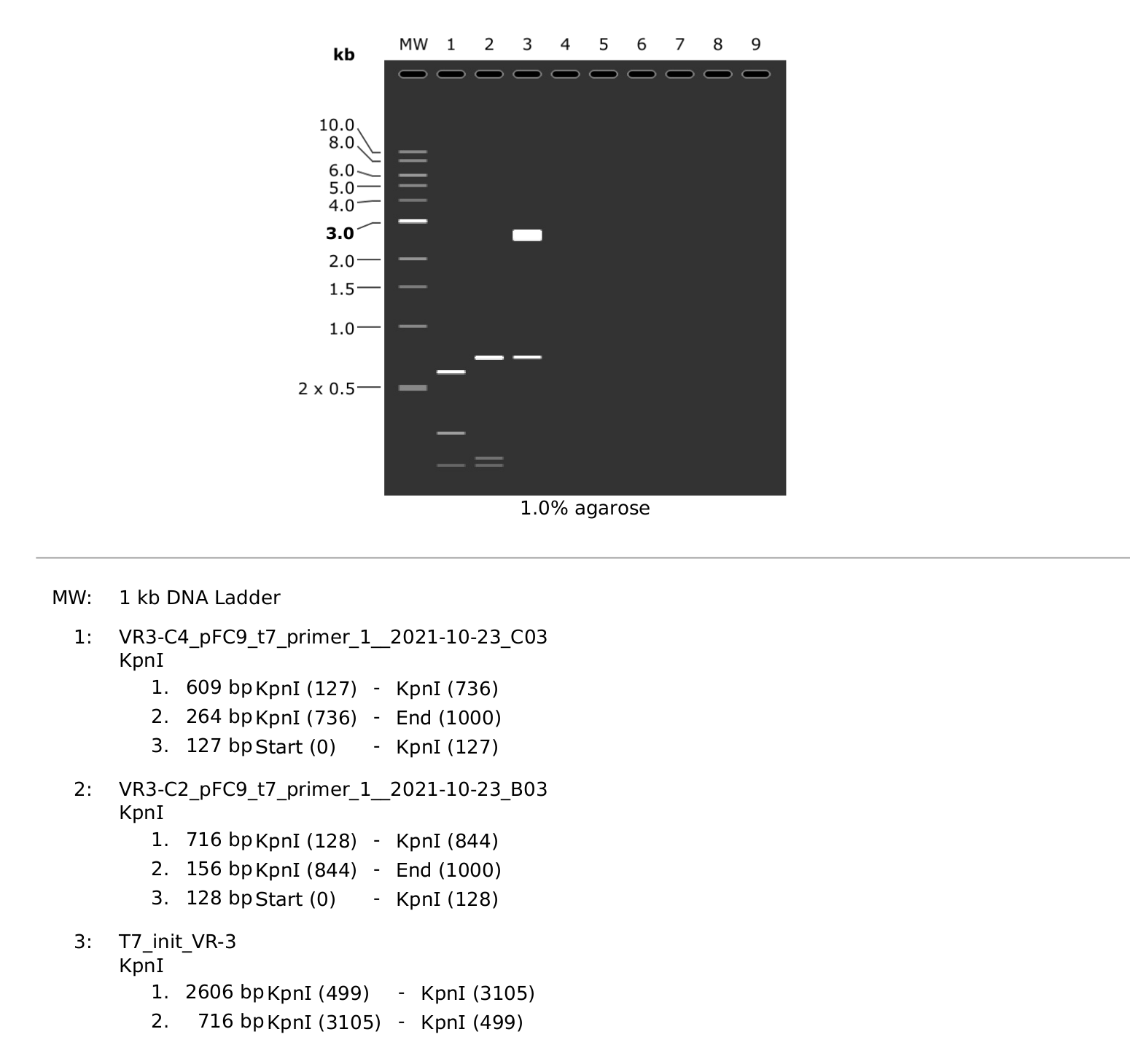
I also sequenced colony 4 from VR-3 because the KpnI digest clearly showed a different band height for C4 compared to other VR-3 samples. The trace looks similar to C2 in its overall quality.
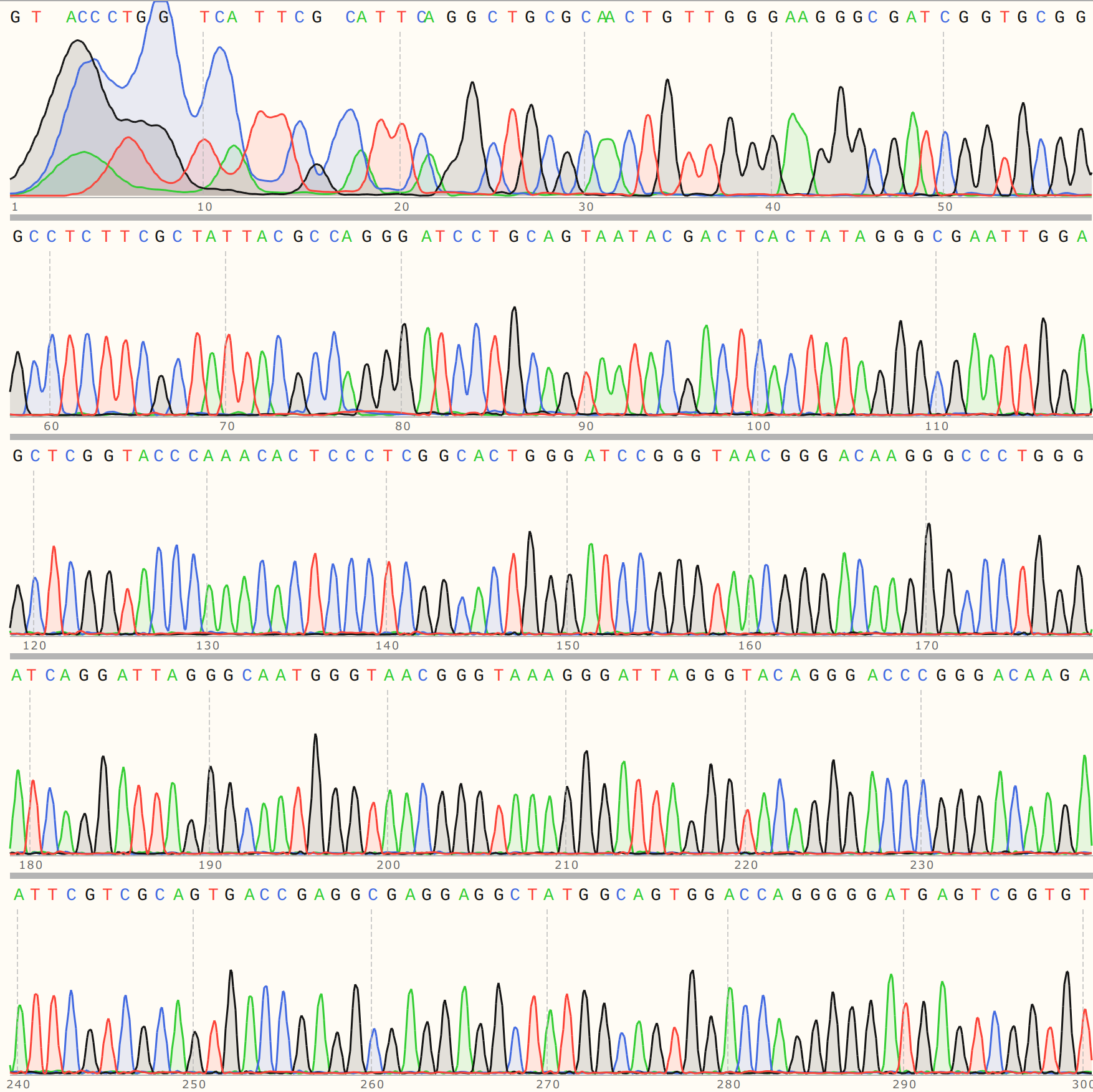
However the detailed alignment shows the most similarity to VR-24, an insert that was not used in any Gibson reactions to date as far as I know.
In fact, looking through the quality control documents I received from Thermo shows that no document was sent for VR-24. Meaning either I missed the shipment or it was never send but it is still showing up in my sequencing results somehow.
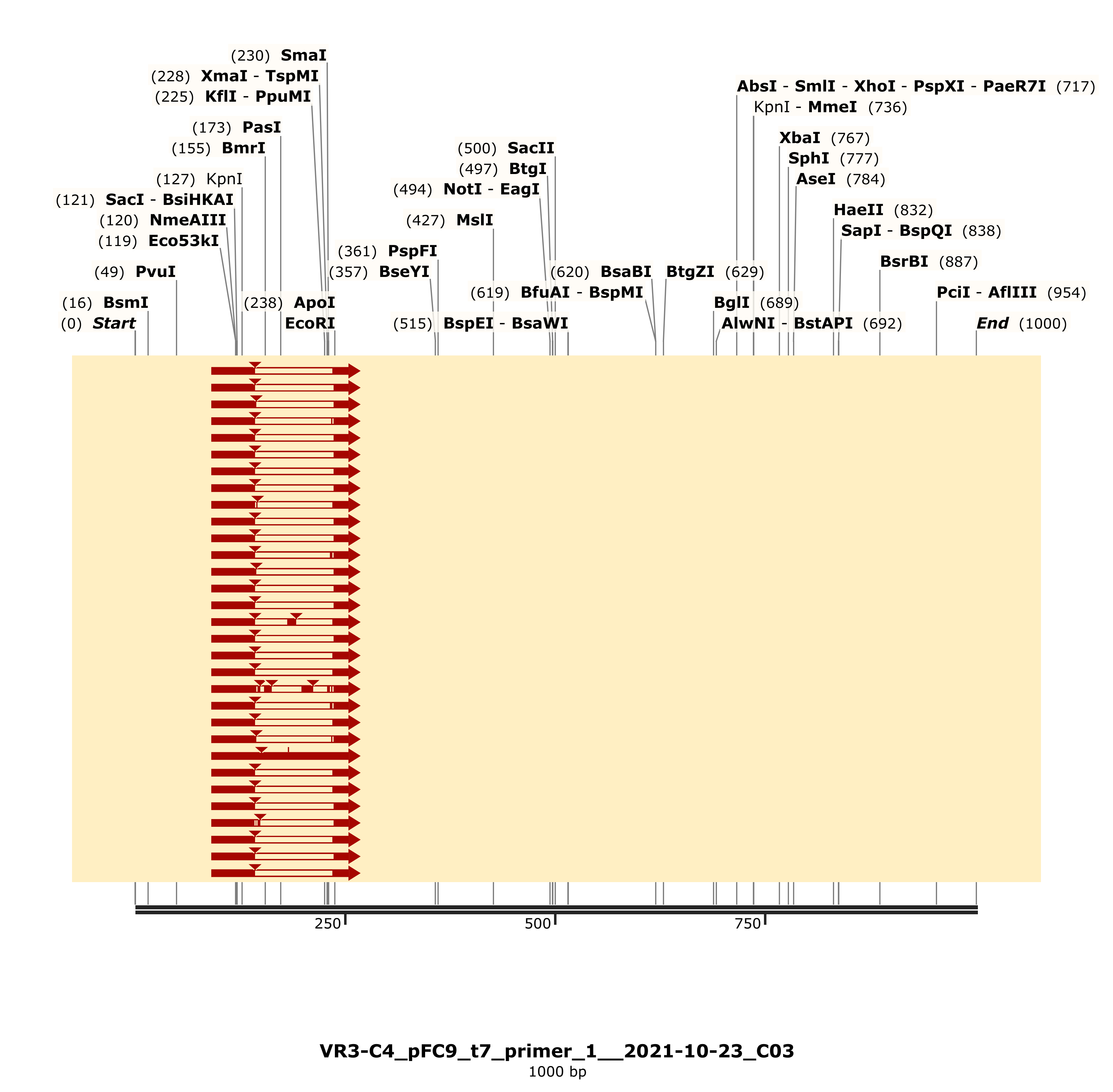
That still leaves the question of why the C4 band was running differently. So I simulated an agarose gel using the Sanger reads and
the simulated VR-3 construct map. This showed different locations
of the KpnI sites in C2 compared to C4 resulting in different fragment
lengths. I am not sure exactly how to explain this currently.
VR-7 #
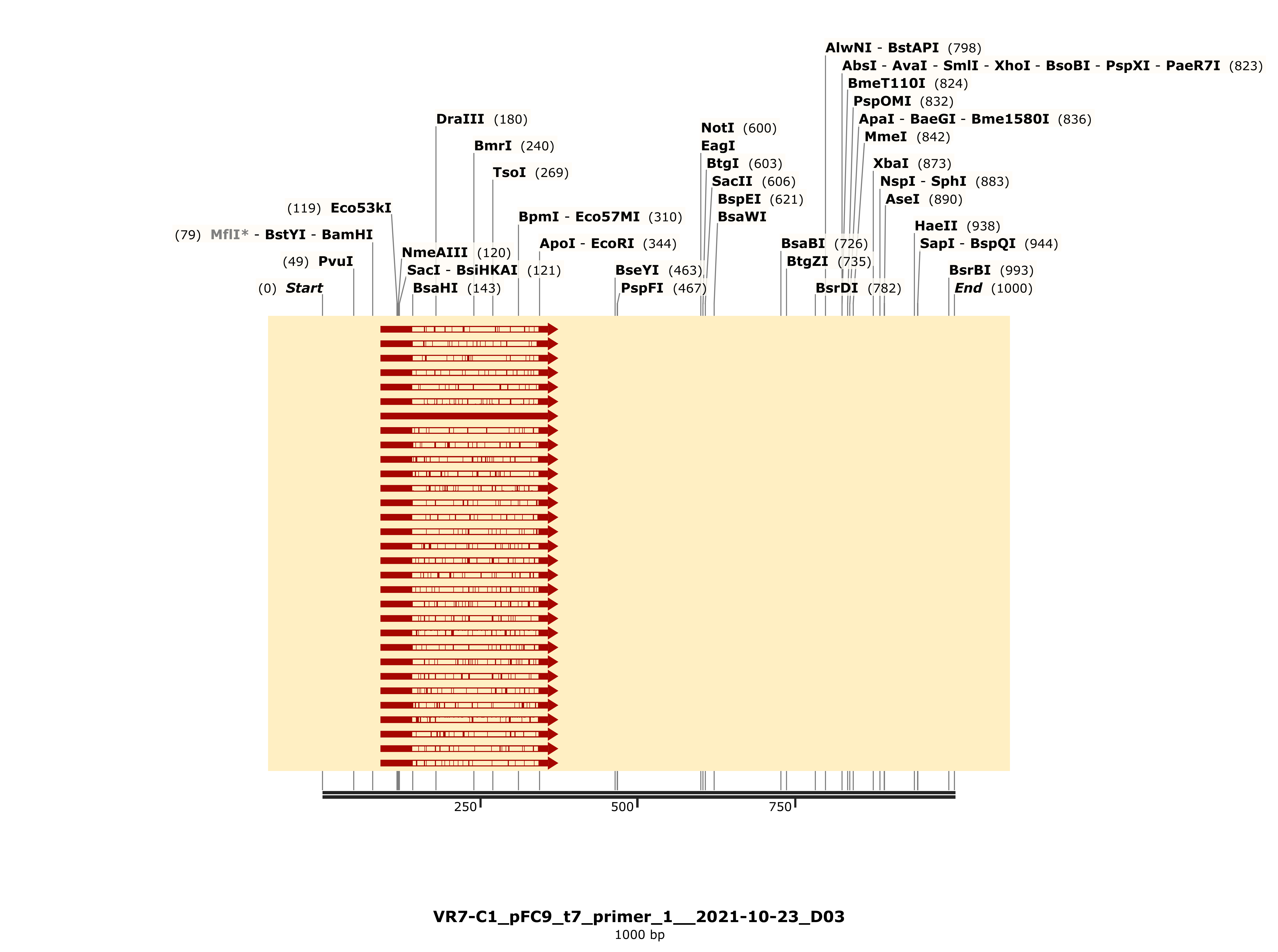
VR-7 might be my favorite of the bunch, traces were clean and BLAST heatmap showed strong alignment to VR-7 reference sequence. Details for C1 are shown below and is representative of results for C2 and 3.
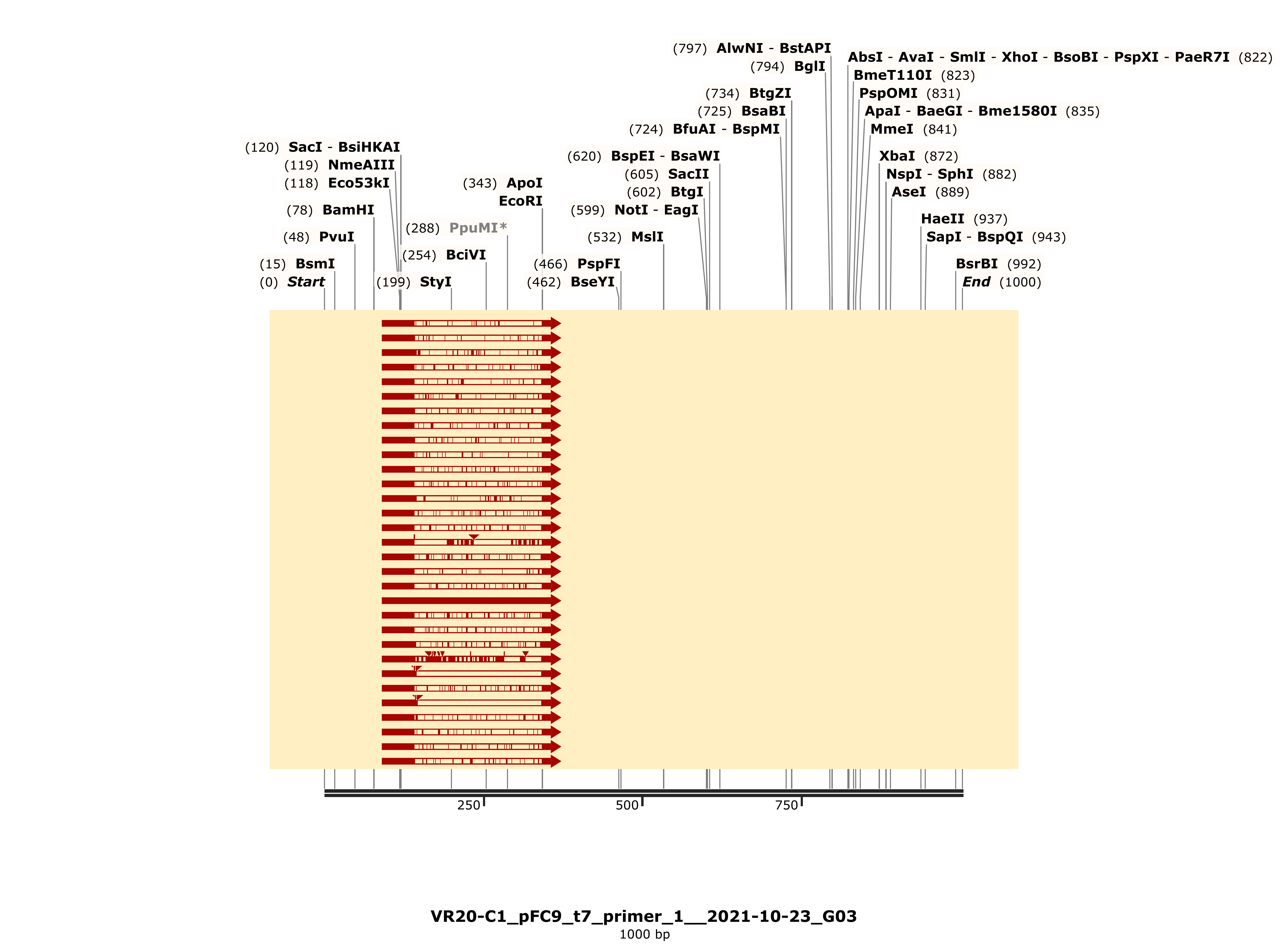
VR-20 #
Got lucky with VR-20, as there seems to be one good sample of the three sent for sequencing.
C1 #
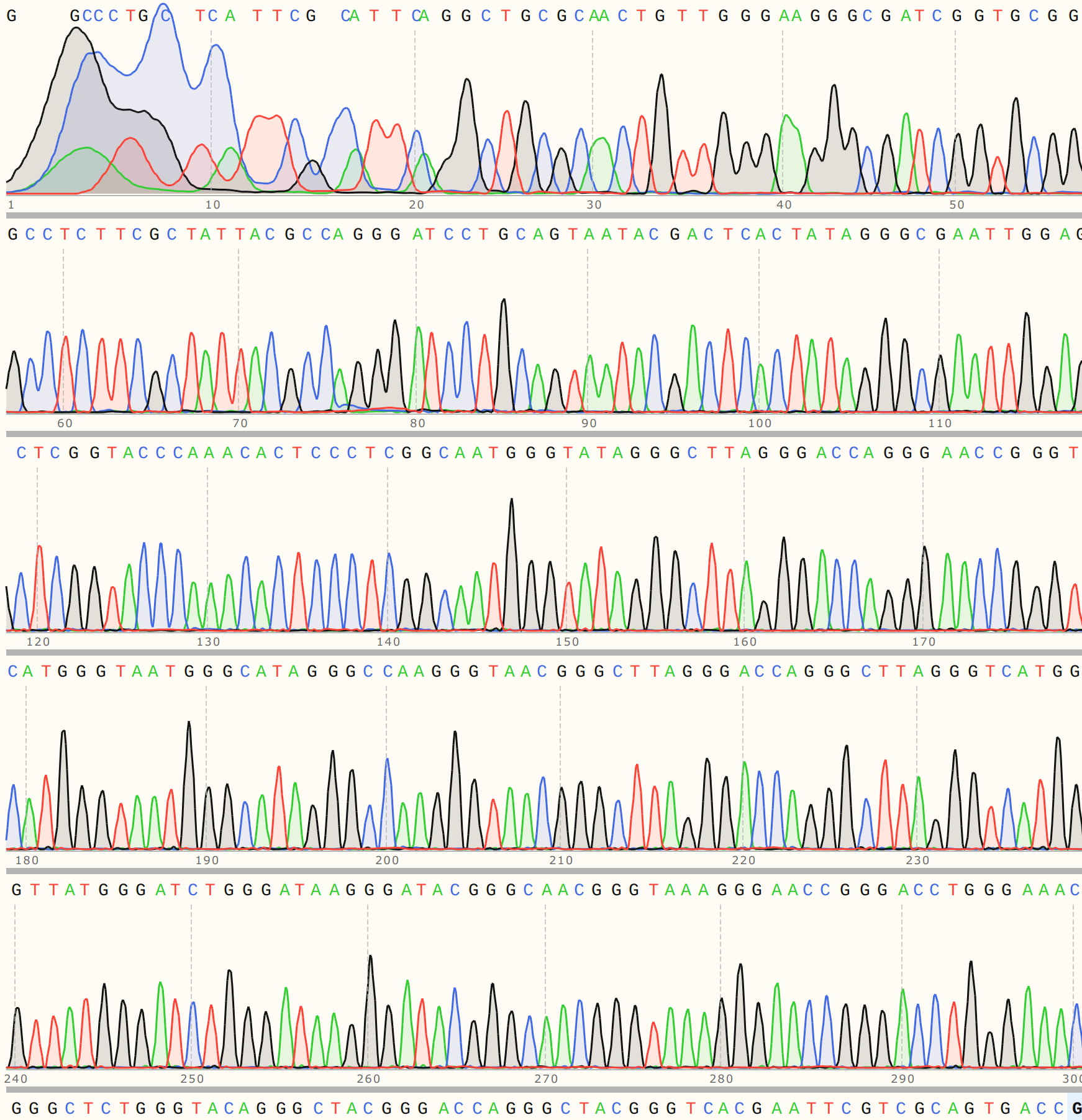
Trace looks strong and shows strong alignment to VR-20 reference sequence.
VR-21 #
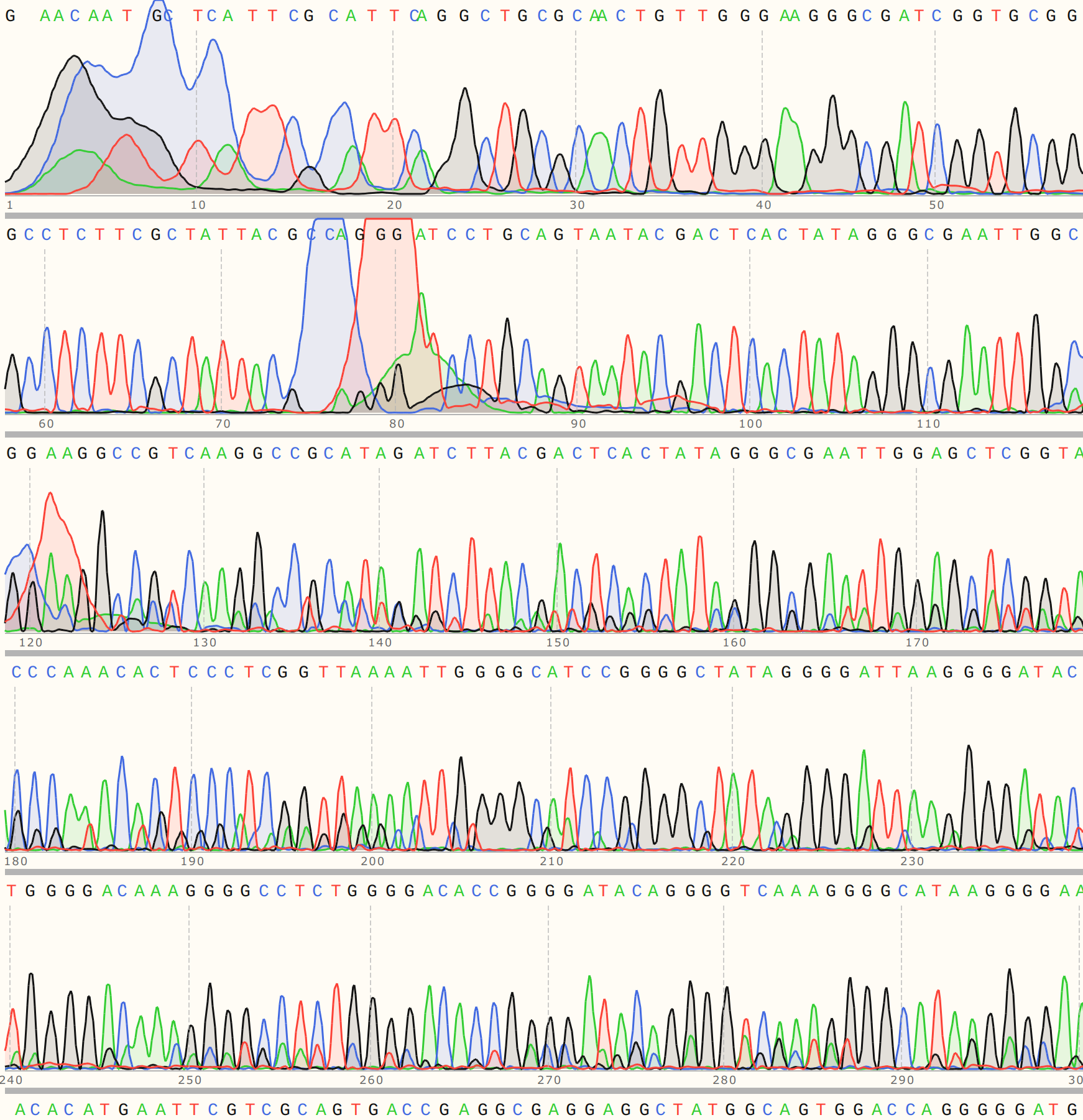
C2 #
Colony 1’s trace was relatively weak, indicating contamination with either another insert of undigested pFC9.
The detailed alignment did show strong alignment to VR-21 reference sequence (with coincidental homology to VR-25).
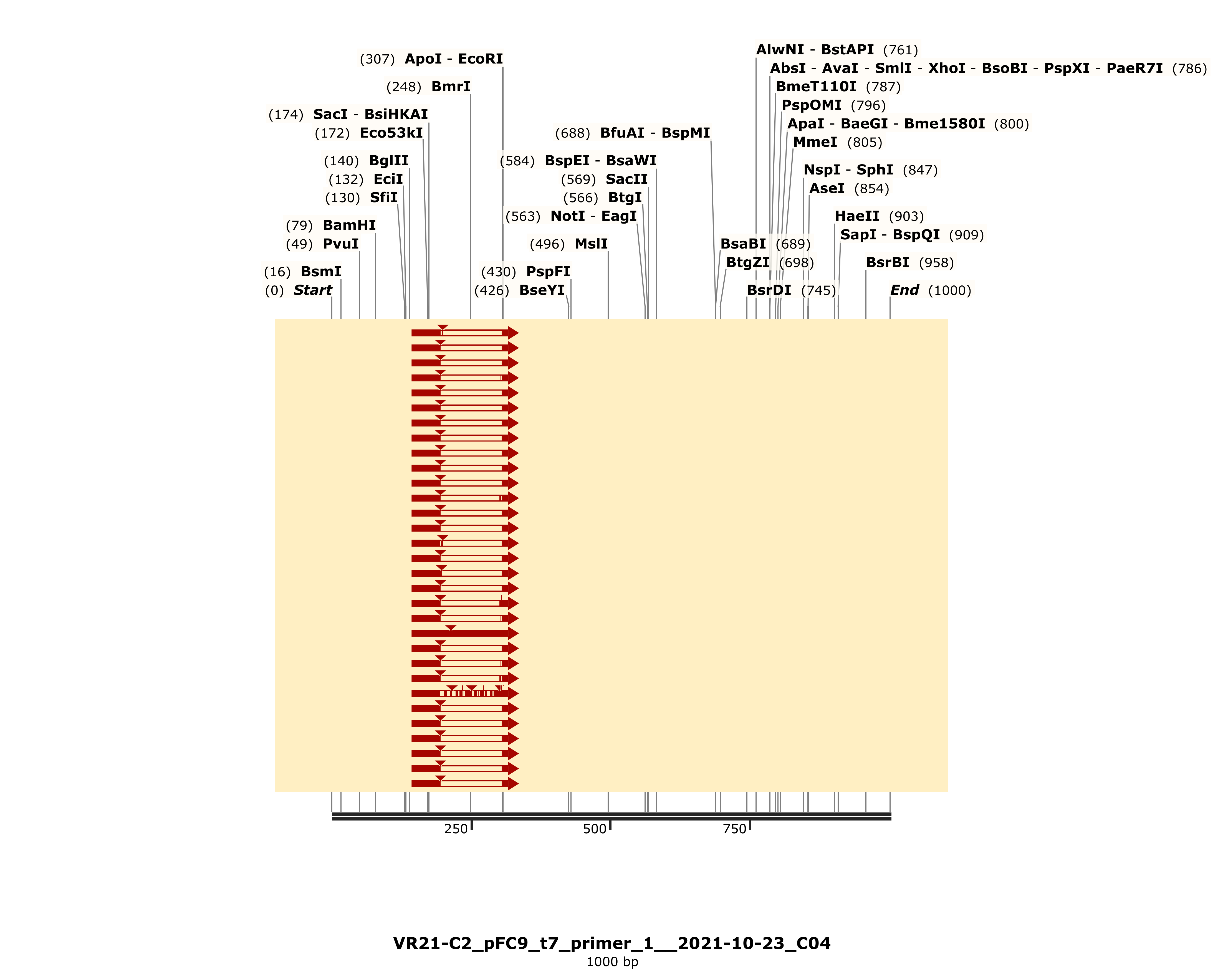
However this alignment is pretty short and includes a gap; coming in at a total length of 164 bp where a full length insert should be 280. This is a bit odd because the variable insert region itself is 200 bp and so each homology arm (which we would expect to align regardless if insert was present or not) only make up 40 bp each of the complete insert. This makes it uncertain to me if it is worth it to try and purify by cloning as it is unclear whether or not the complete insert is present.
I saw a similar pattern in sequence alignments for colonies 5 and 6 but the traces looked cleaner (trace for VR-21-C5 is shown below).
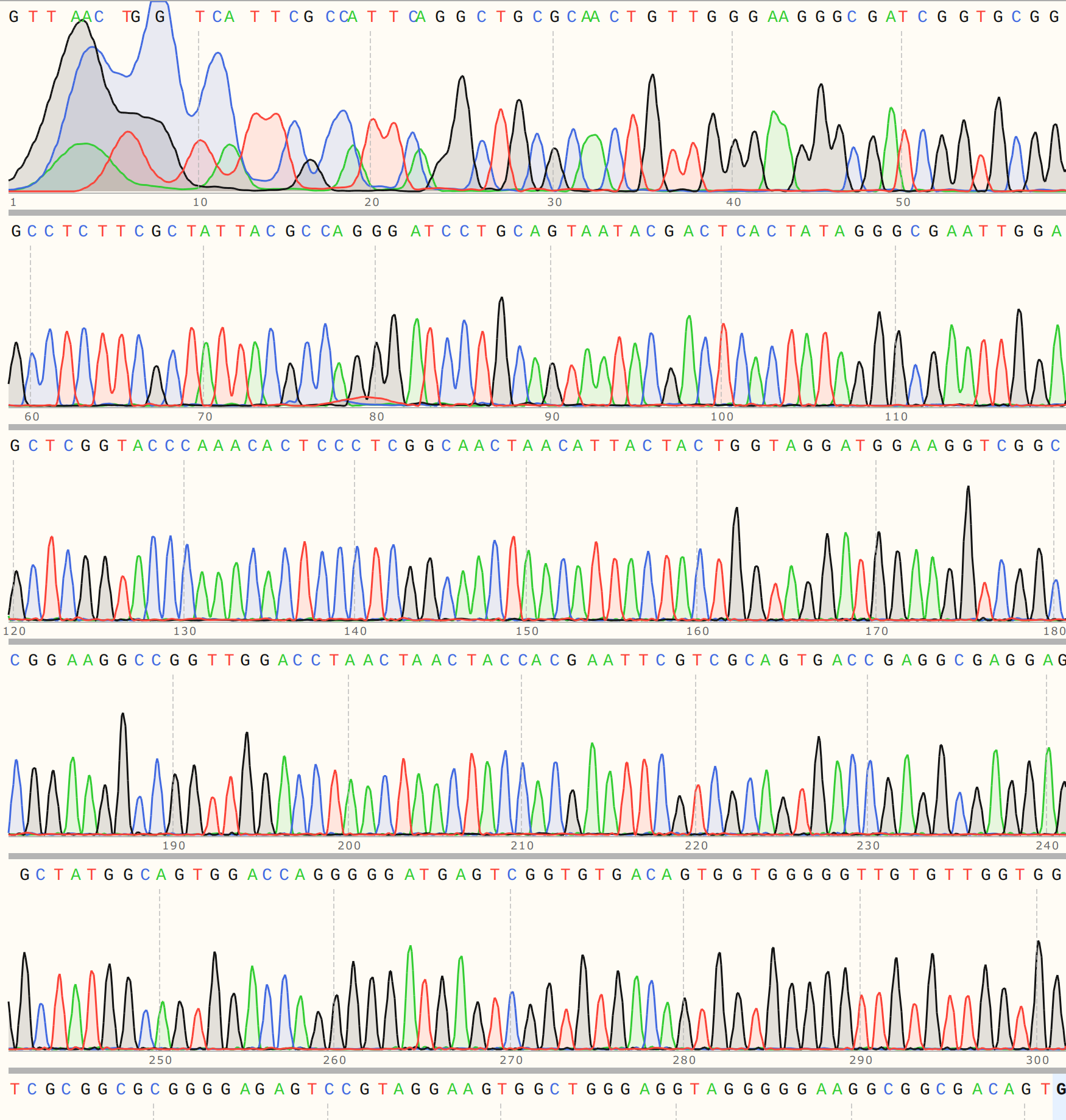
This seems to be pointing towards the presence of a truncated insert.
VR-30 #
C1 #
Colony one showed strong alignment to the VR-30 reference sequence and trace looked strong.
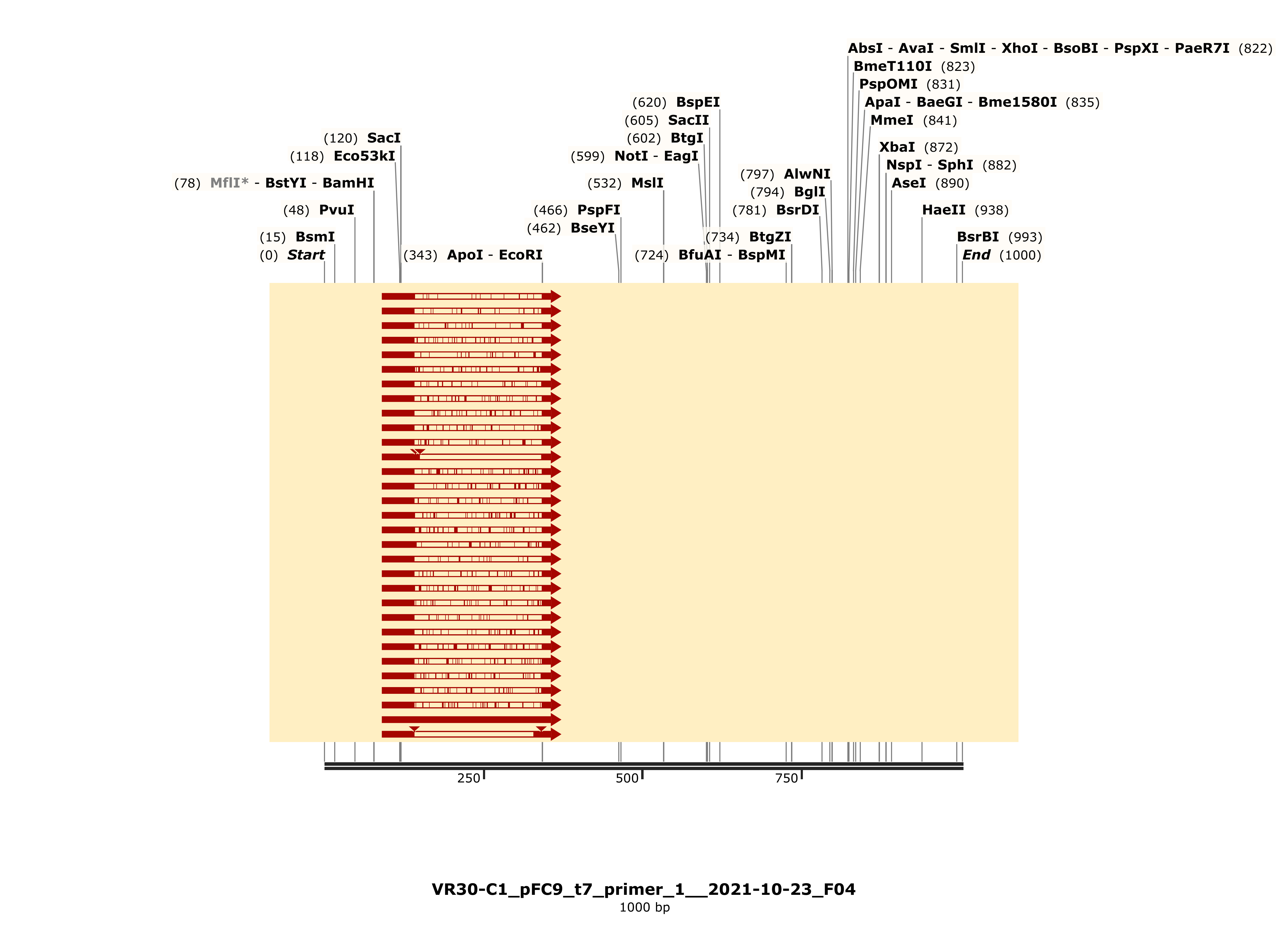
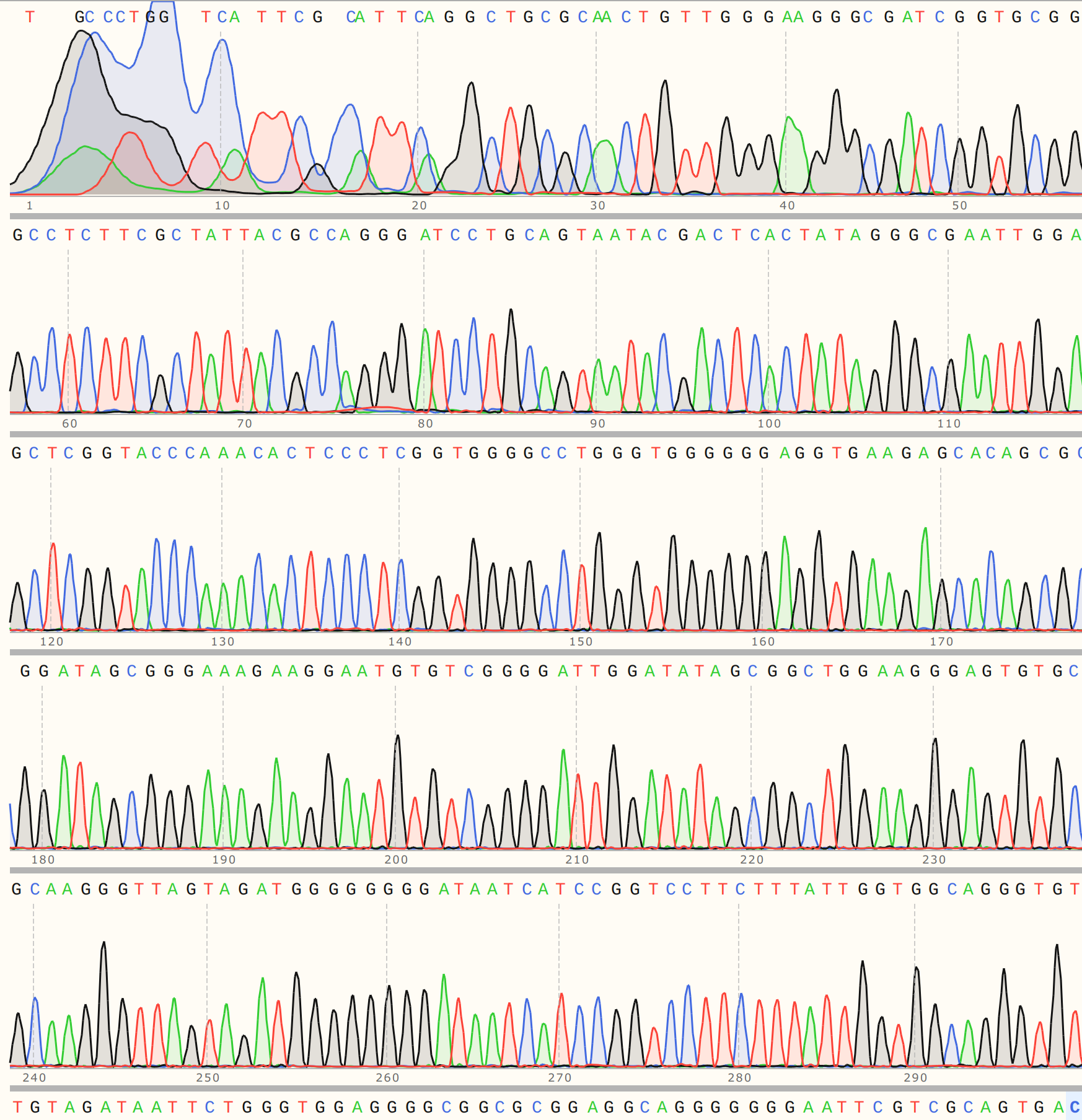
C3 #
Colony three aligned strongly to VR-3 reference sequence VR-3 with a strong trace to go along with it.
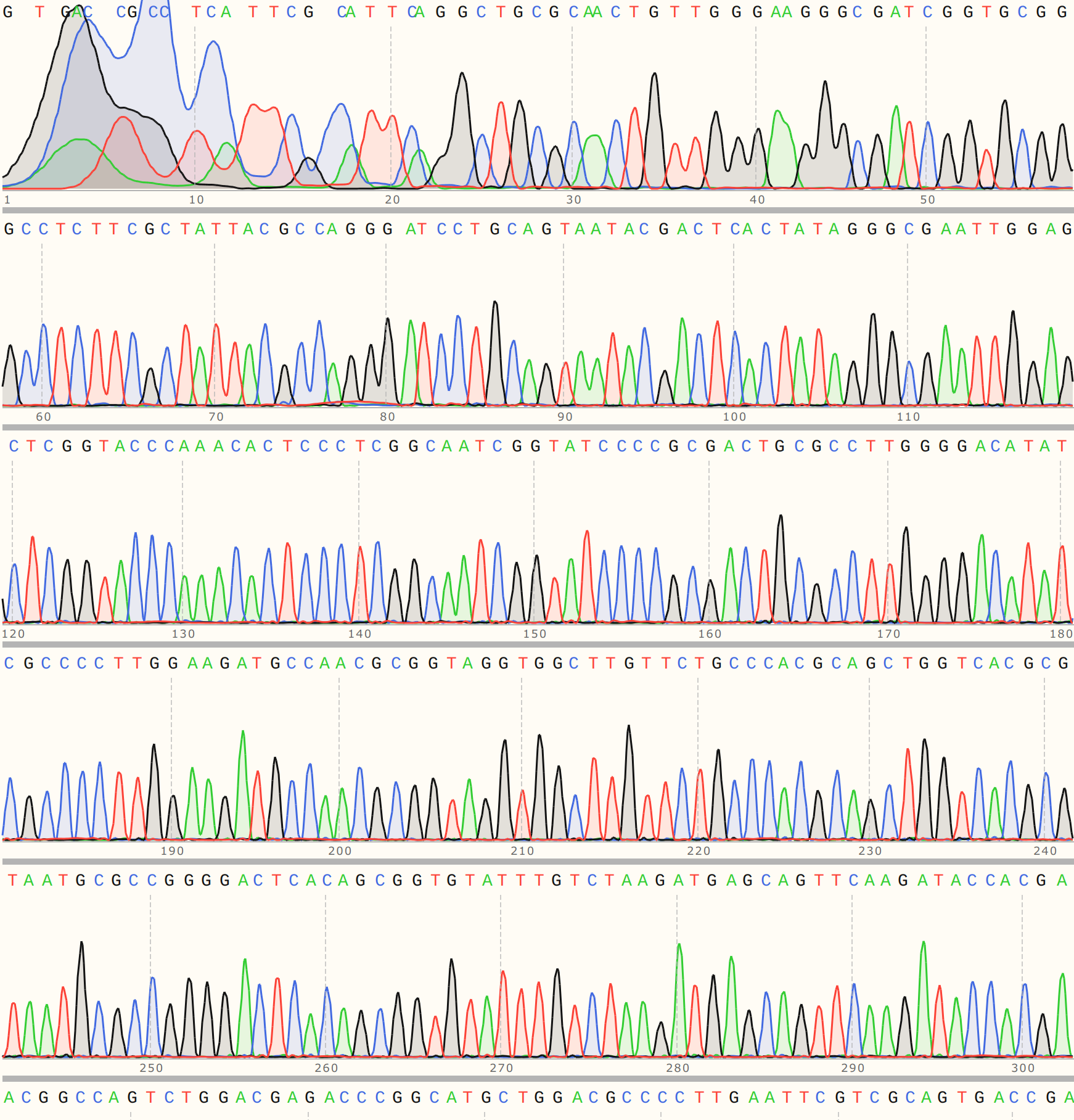
This points towards contamination in VR-30 samples with VR-3 at some point along the reagent handling chain. Difficult to say exactly where at this point.
VR-31 #
All VR-31 reads were very short compared to other samples. Read lengths are shown in the plot below, no pruning my base quality was done.
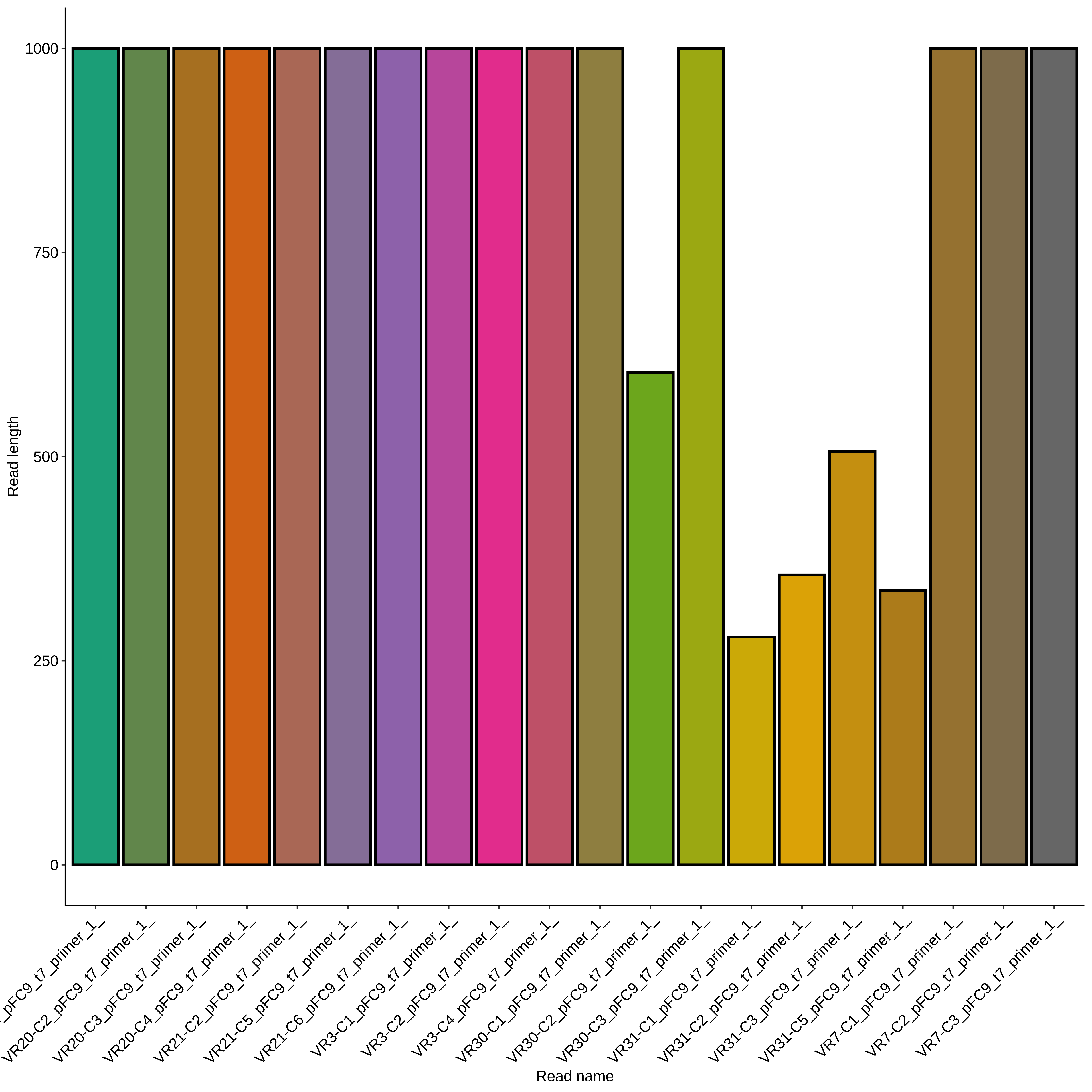
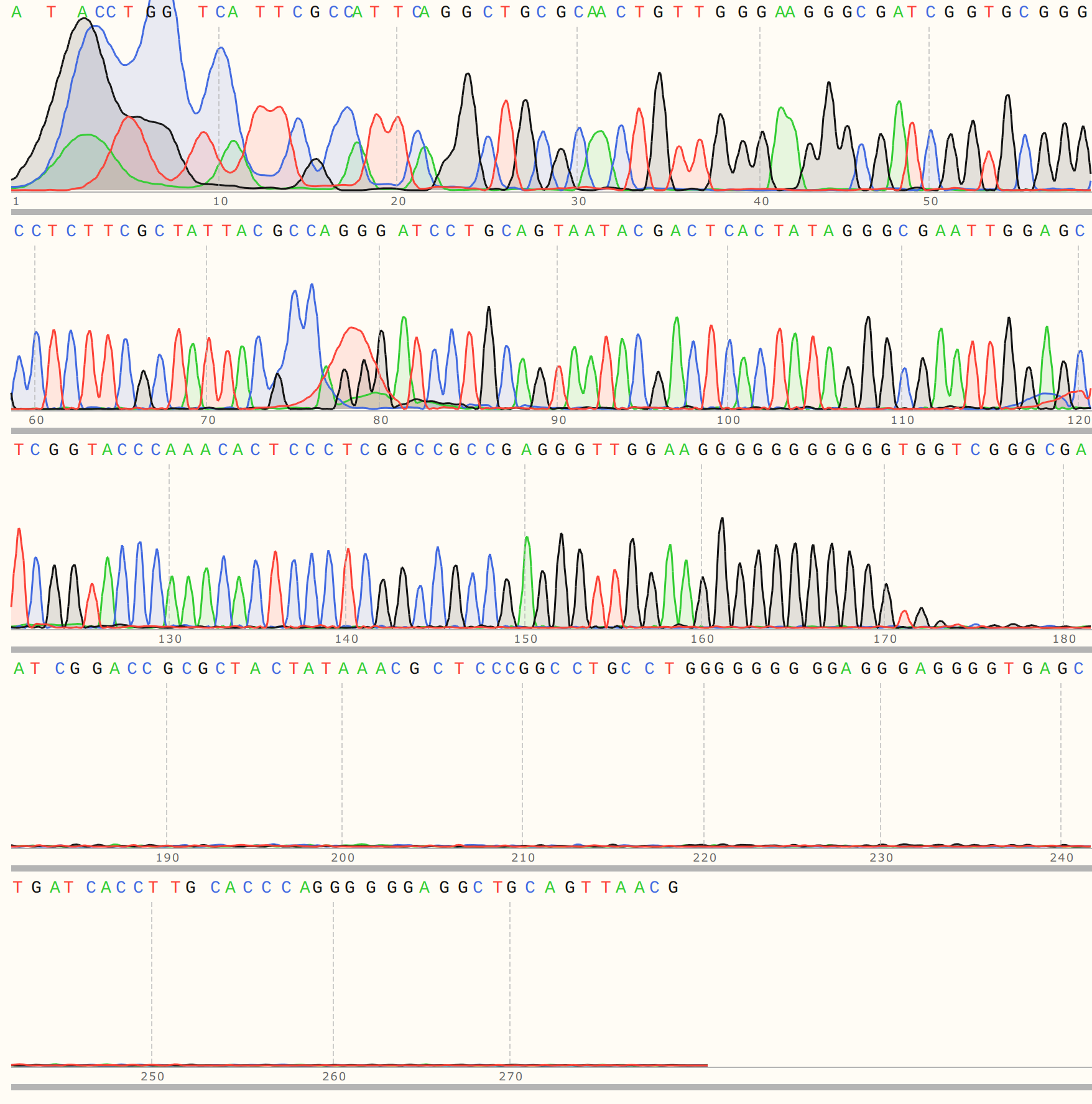
All VR-31 reads showed clean but short traces. Trace for colony 1 shown below (representative of C2, 3 and 5).
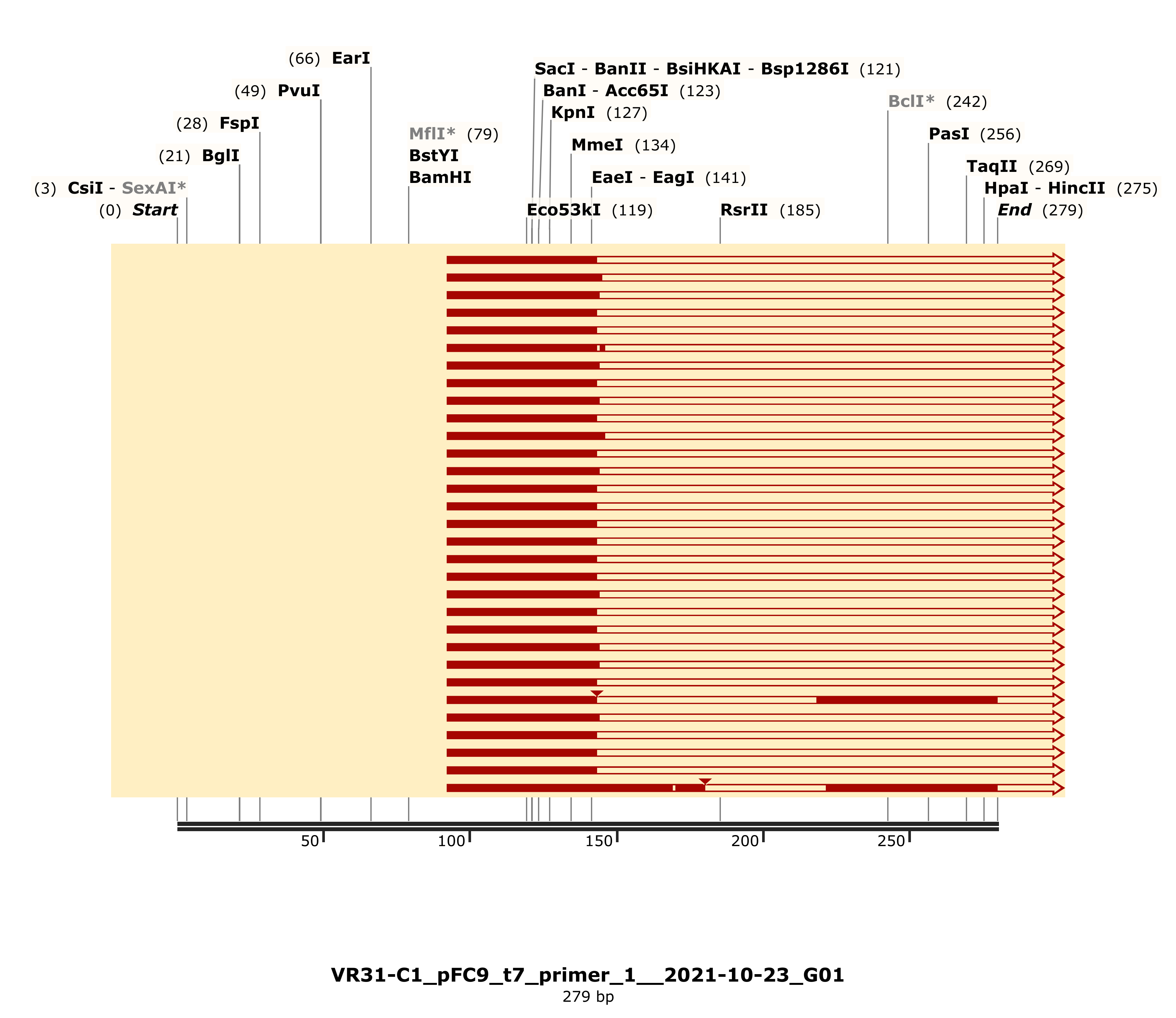
Alignments showed large gaps. Detailed alignment for C1 shown but largely representative of other VR-31 samples.
This one may be the most confusing. The primer was able to bind and sequencing was able to start but quickly died. And not just died as in calls became random, there were no calls at all for any of these samples. These samples still digested with KpnI though.
The takeaway #
Given analysis above table below is what I am thinking for next steps.
| Insert | Action |
|---|---|
| VR-3 | Purification by cloning: C1 |
| VR-7 | None. Relabel VR-7-C1 as pFC9VR7 |
| VR-20 | None. Relabel VR-20-C1 as pFC9VR20 |
| VR-21 | Redo fragment purification and Gibson assembly |
| VR-30 | None. Relabel VR-30-C1 as pFC9VR30 |
| VR-31 | Talk to Fred. Likely redo fragment purification and Gibson assembly. |