Enzyme and reagent validation #
Link to spreadsheet describing validation results
T7 Pol from Fraser lab #
1/5/22 #
Tada’s tested T7 from Fraser lab via IVT of pFC9 and serial dilution of the provided T7. He also aliquoted the 1 ml volume received into 5 0.5 ml tubes.
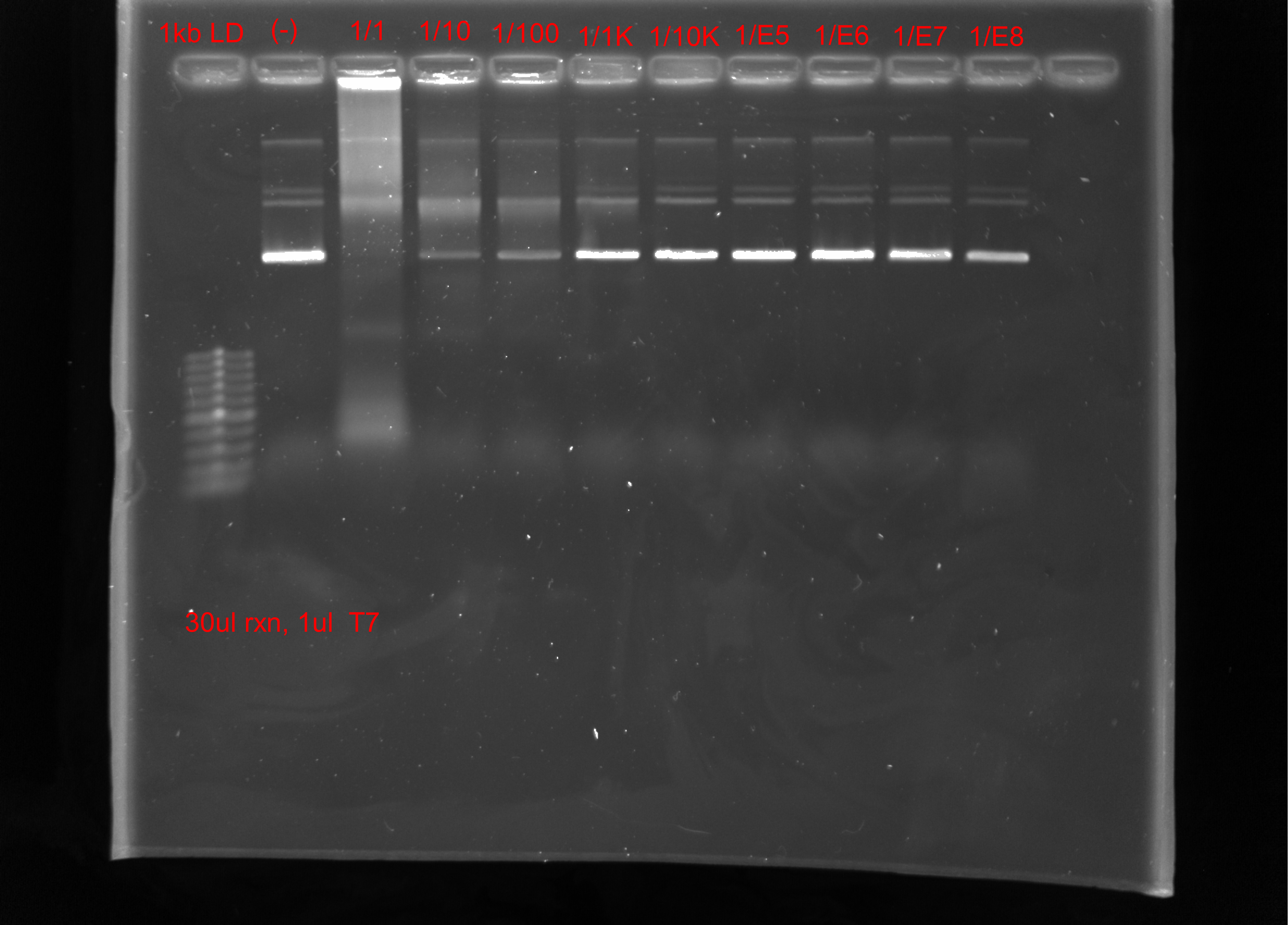
Based on these results, T7 should be used as is, 1/10 dilution already greatly decreases the apparent band shift compared to control. 1/1000 dilution looks basically the same as the control.
NEB Topoisomerase validation #
1/10/22 #
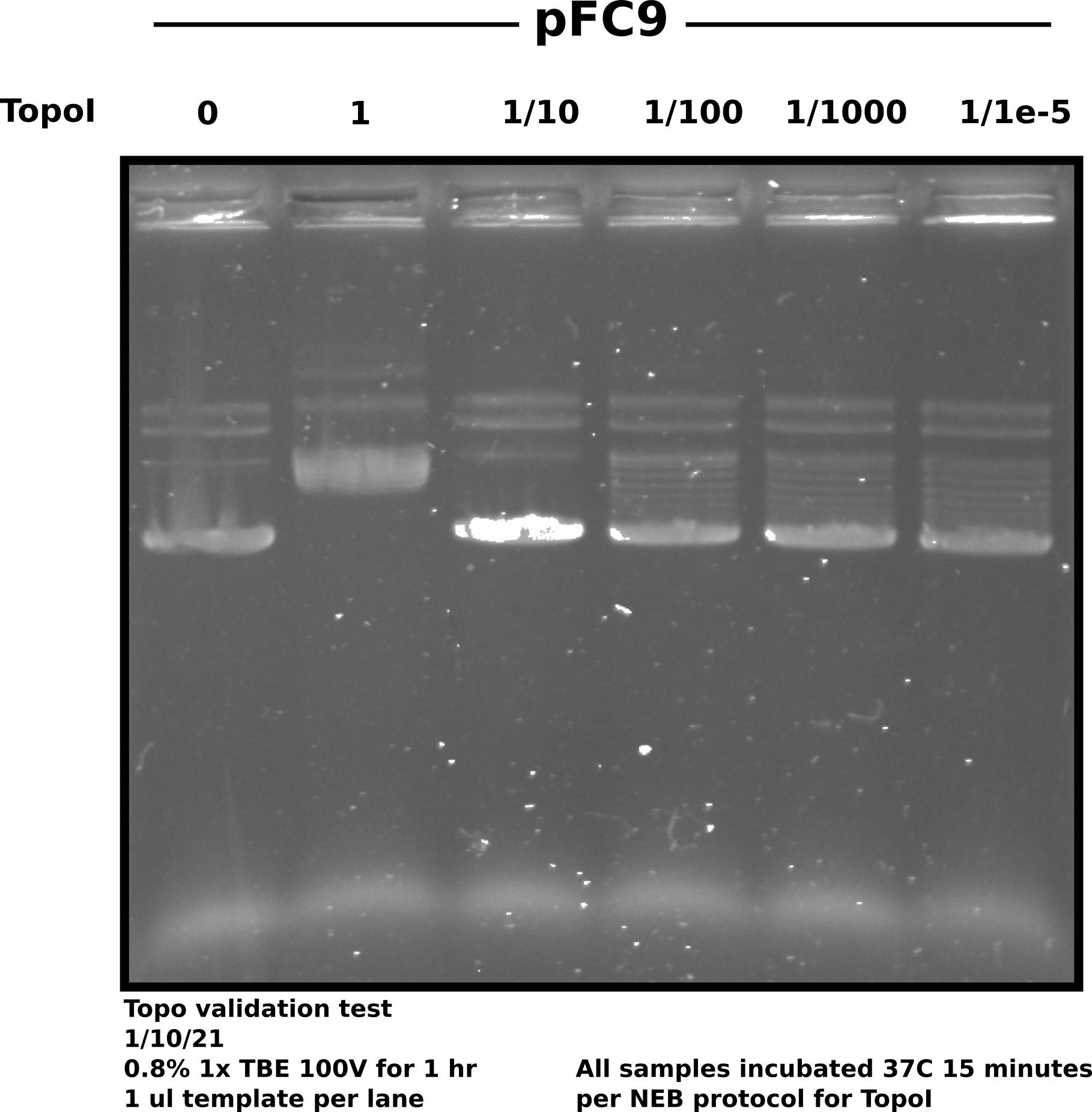
Validating the activity of TopoI that came in today using pFC9 as substrate for relaxation.
Protocol #
| Sample | Template DNA | 10x rCutSmart | Template DNA mass (ng) | Template DNA volume (ul) | TopoI volume (ul) | TopoI concentration | npH20 Volume (ul) | Incubation temp (C) | Incubation time (mins) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | pFC9 | 2 | 600 | 2 | 0 | NA | 15 | 37 | 15 |
| 2 | pFC9 | 2 | 600 | 2 | 0 | 1 | 15 | 37 | 15 |
| 3 | pFC9 | 2 | 600 | 2 | 0 | 0.1 | 15 | 37 | 15 |
| 4 | pFC9 | 2 | 600 | 2 | 0 | 0.01 | 15 | 37 | 15 |
| 5 | pFC9 | 2 | 600 | 2 | 0 | 0.001 | 15 | 37 | 15 |
| 6 | pFC9 | 2 | 600 | 2 | 0 | 0.0001 | 15 | 37 | 15 |
Topo concentrations
1 ul of Topo was used in sample 2 (Topo concentration of 1). The result of Topo treated samples were from a serial dilution of 1 ul Topo in 9 ul 1x CutSmart buffer. 1 ul of each dilution was used in each sample.
Prepared samples as described by table above. After 15 minute incubation
ran samples on 0.8% 1x TBE in 1x TBE buffer @ 100V for 45 minutes.
Post-stained for 30 mins with 2 ul EtBr / 100 ml TBE buffer.
Used new rCutSmart buffer cat number B6004S lot number 10123108 for all
Topo reactions.
Results #
NEB Nt.BspQI #
1/11/21 #
Nt.BspQI is a nicking endonuclease. Purchased from NEB cat number R0644S. Lot number 10117885 opened 1/11/21.
Protocol #
| Sample | Template | Template volume (ul) | Buffer r3.1 volume (ul) | Enzyme (ul) | npH20 |
|---|---|---|---|---|---|
| 1 | pFC9VR14 | 1 | 5 | 1 | 45 |
| 2 | pFC9VR14 | 1 | 5 | 1 | 44 |
| 3 | pFC9VR14 | 10 | 5 | 1 | 34 |
| 4 | pFC9VR14 | 30 | 5 | 1 | 14 |
All samples incubated 1 hour 50C then heat kill at 80C for 20 mins per NEB provided protocol. In addition to testing nicking activity I am also testing how much 1 ul of enzyme can nick in the prescribed reaction time period.
Agarose gel #
In order to compensate for higher concentrations of VR14 in sample 3 and 4 loaded smaller volumes that would be equivalent to loading 25 ul of samples 1 and 2; shown in table below. Difference in volume was made up with npH20.
| Sample | Total Volume (ul) | Total loading volume (ul) | Sample volume loaded (ul) | npH20 | npH20 |
|---|---|---|---|---|---|
| 1 | 50 | 25 | 25 | 0 | 45 |
| 2 | 50 | 25 | 25 | 0 | 44 |
| 3 | 50 | 25 | 2.5 | 22.5 | 34 |
| 4 | 50 | 25 | 0.833 | 24.167 | 14 |

Based on gel Nt.BspQI is a very efficient nickase and standard reaction setup can be used for a large amount of template DNA.
Topogen E. coli DNA Gyrase #
2/9/22 #
Received Topogen E. coli DNA gyrase and placed into -80C freezer on arrival. I then aliquoted into 2 ul (small I know but there are only 10 ul total) after thawing on ice in the cold room. Stored aliquots back in the -80.
Reagent details #
- Lot number: 22JA25
- Concentration: 10 u/ul
2/10/22 #
Validation experimental protocol #
In order to validate Gyrase activity using it to attempt to repair nicked pFC9VR14 sample. First I used Nt.BspQI nicking enzyme to convert any remaining supercoiled molecules to nicked circular plasmids. Then I plan to use DNA taq ligase to repair nicks produced by Bt.BspQI. This will produce a homogenous population of relaxed circular plasmids. I then will conduct a time course experiment with DNA gyrase to determine activity and incubation times for future nicked plasmid repair. Spreadsheet describing reagent details and volumes is available at this link.
Nt.BspQI digestion #
Prepared samples according to Nt.BspQI reaction conditions and incubated at 50C for 1 hour followed by 80C for 20 minutes to head kill enzyme.
Reaction conditions #
| npH20 | r3.1 10x Buffer | Plasmid | Plasmid volume (ul) | Nt.BspQI (ul) | Reaction volume (ul) |
|---|---|---|---|---|---|
| 8.389113771 | 2.5 | pFC9-T7-init-VR14-2-midi-D | 13.11088623 | 1 | 25 |
Taq ligase treatment #
Transfered 16.6 ul of Nt.BspQI treated template into new reaction with Taq ligase described by reaction conditions table below. Incubated sample for 30 mins at 45C in the thermocycler.
Reaction conditions #
| Volume nicked substrate | npH20 | Taq Ligase 10x Buffer | Total volume (ul) | Taq ligase (ul) | Reaction volume (ul) |
|---|---|---|---|---|---|
| 16.667 | 26.333 | 5 | 50 | 2 | 25 |
After completing Taq ligase reaction I preformed standard phenol-chloroform EtOH purification to remove Nt.BspQI and Taq ligase. Re-suspended with 50 ul of 10 mM Tris HCl.
DNA Gyrase treatment #
After completing Taq ligase treatment and precipitation I transferred 24.9 ul of nicked and ligated sample to a new reaction. While working in the cold room and on ice, I diluted DNA gyrase to 5 units / ul (2 fold) from aliquot 1 using the Topogen provided dilution buffer. I then assembled all reaction components (enzyme last) into one 0.5 ul tube. I then distributed 5 ul of master mix into separate individual PCR tubes. I then placed all samples into the thermocycler and began incubation at 37C. At each designed time point (shown in table below) I removed the respective sample, placed on ice and added 1 ul purple loading dye. I continued this for all samples.
Reaction conditions #
| Volume ligated substrate | npH20 | 5x Topogen Gyrase buffer | Gyrase (5u/ul) ul | Total Volume | |
|---|---|---|---|---|---|
| 24.99950001 | 13.00049999 | 10 | 2 | 50 |
Time points assayed #
| Gyrase Rxn time points (min) | Aliquot volume (ul) |
|---|---|
| 10 | 5 |
| 15 | 5 |
| 20 | 5 |
| 30 | 5 |
| 40 | 5 |
| 50 | 5 |
| 60 | 5 |
| 80 | 5 |
| 120 | 5 |
Analysis of DNA gyrase activity by agarose gel #
I prepared a 0.8% 1x TAE SeaPlaque agarose gel without EtBr. Ran gel for 5 hours time at 60V at room temperature. After running gel I post-stained with EtBr at a concentration of 5 ul / 100 ml TAE for 15 minutes and then destained in TAE for 10 minutes. ~ 100 ng of sample per lane. While loading the gel I noticed that the 60 minute Gyrase incubation timepoint and noticeably less volume then other samples; this may have been due to pipetting error (bubble maybe?).
DNA gyrase test on repaired nicked plasmids #
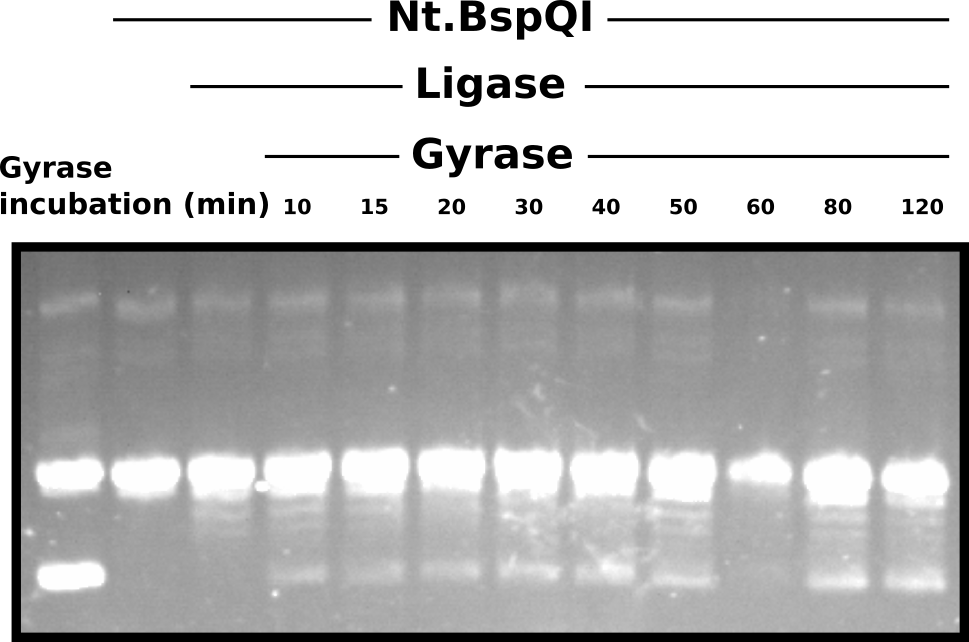
Clearly Nt.BspQI treatment nicked all molecules, converting template to virtually 100% nicked. While it is difficult to definitively determine if the ligation reaction was successful then presence of small sub-bands below the large nicked band may indicate some degree of success. However, Gyrase feels to have failed to supercoil most of the “repaired” plasmid. In my mind there are three possible reasons for this.
- Gyrase is in-active
- Much more DNA was added then I thought I was adding due to in-accurate concentration measurement. So the expected amount was converted to supercoiled substrate but it appears to be small compared to the large amount of DNA present in the sample.
- Ligation reaction did not successful repair all of the template present
(this explanation is not mutually exclusive from
2) and so there was little substrate that Gyrase could actually converted to supercoiled as catalysis on nicked substrate would still result in a relaxed molecule.
In order to diagnosis I will preform a follow up experiment that tests Gyrase activity independently of nick repair by relaxing pFC9 with Topoisomerase, proteaseK digesting and phenol-chloroform precipitation and then incubating with Gyrase for 1 hour.
2/11/22 #
Positive control using Topoisomerase I relaxes substrate #
In order to test activity of Gyrase without requiring a ligation reaction I used NEB E. coli Topoisomerase I to relax pFC9. I then treated this relaxed substrate with Topogen gyrase.
Protocol #
- Treat 1 ug pFC9 (3.33 ul) with TopoI according to NEB provided protocol.
- Digest sample with 1 ul proteaseK at room temperature for 10 minutes.
- Preform standard Phenol-chloroform EtOH precipitation. Re-suspend samples in 10 ul of 10 mM Tris HCl.
- Add 9 ul TopoI treated template to 30 ul npH20, 10 ul Topogen 5x Assay buffer. Save the remaining 1 ul for later analysis on agarose gel.
- Working in the cold room, thaw aliquote 1 dilution 1 (5 units / ul) of DNA gyrase on ice.
- Add 1 ul (5 units) of DNA gyrase to sample. Keep sample on ice and return to lab. Place sample in thermocycler at 37C for 1 hour.
- After 1 hour aliqote 5.55 ul to a new PCR tube. Repeat this step at 1.5 and 2 hour time points.
- Analyze samples on agarose gel no EtBr. Post-stain gel after finished running.
2/15/22 #
Completed the protocol described above with pFC9. Agarose gel of samples is shown in the image below.
Topoisomerase I treatment followed by DNA gyrase #
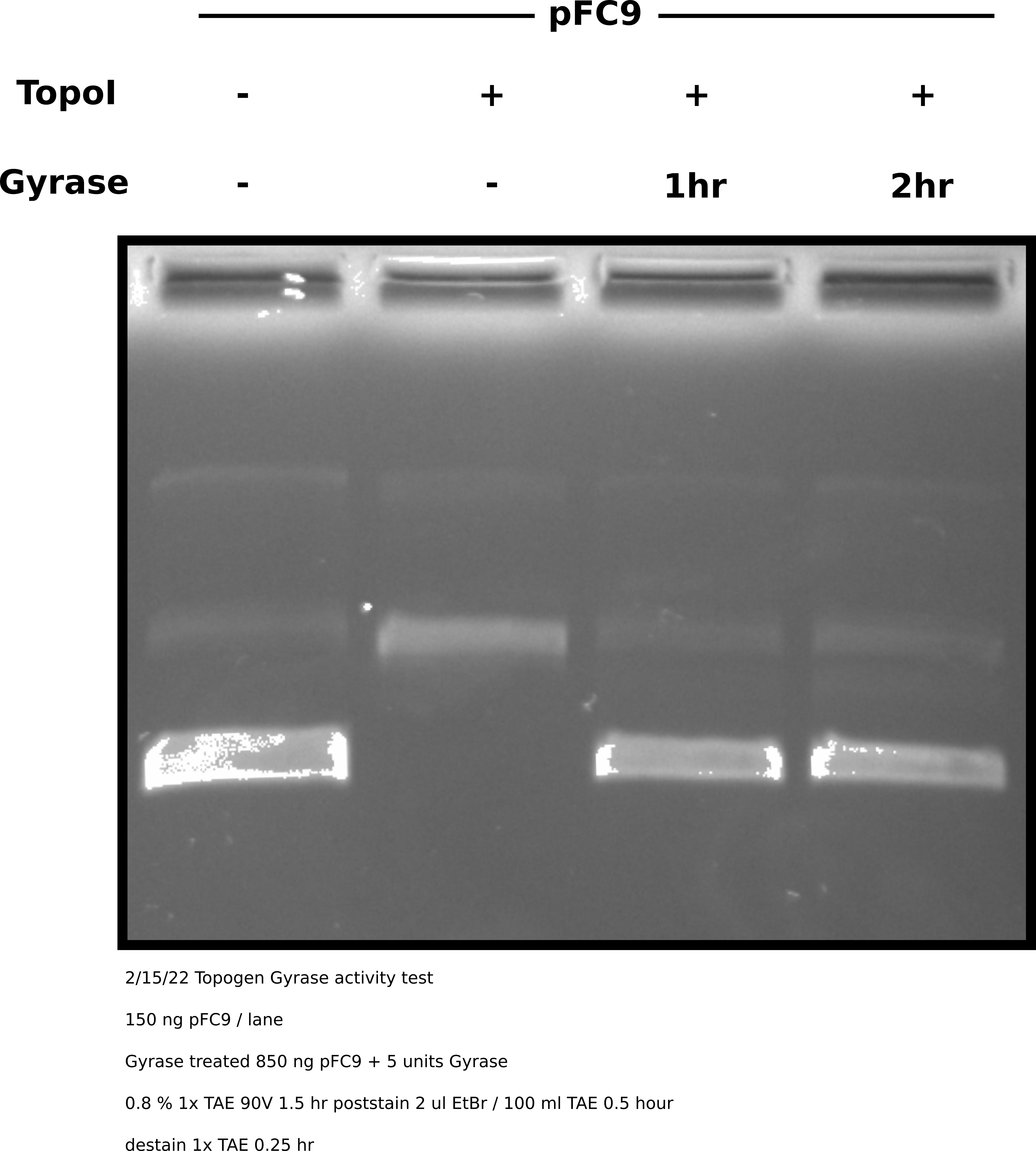
Gyrase clearly shows full activity and is able to almost completely supercoil the relaxed plasmid. This indicates that its failure to do so after treating nicked plasmids with Nt.BspQI was due to nicks that Taq ligase failed to repair. Therefore Gyrase is active but nick repair is not a viable option without optimization. Even then it may not be due to the high cost of Gyrase enzyme.