Test for possible nuclease activity in RNAse A enzyme stock #
Follow up to 6-1-21 test using plasmids pFC11 and pFC44. Also noted that running pretty low on stock of pFC14 and 17. Need to make more soon.
Protocol #
Plasmid concentrations table.
| Plasmid | DNA Concentration (ng / ul) |
|---|---|
| pFC11 | 1080 |
| pFC44 | 1165 |
DNA master mix recipe.
| Plasmid | H20 (ul) | DNA (ul) | Total Volume (ul) |
|---|---|---|---|
| pFC11 | 88.34 | 1.66 | 90 |
| pFC44 | 88.46 | 1.54 | 90 |
Stock RNase A (1mg/ml) was diluted 1:100.
Master mixes described above were prepared and then separated into 8 aliquots per sample. 8 samples were divided into 4 increasingly long durations of incubation at 37C for 15, 30, 60 and 120 minutes. Samples were added to the thermocycler in reverse order by duration of incubation (120 mins first) and 4 ul of RNAse A was addded to treated samples while 4 ul npH20 was added to controls immediatly before placing samples into the thermocycler.
After all samples completed incubation I had to give a presentation in one of my classes and so there was a delay of about 1hr between the last sample finishing incubation and actually running samples on agarose gel.
Samples ran on 0.7% agarose gel for 45 mins at 90 V in TAE with 12 ul of EtBr in both agarose mix and running buffer.
Results #
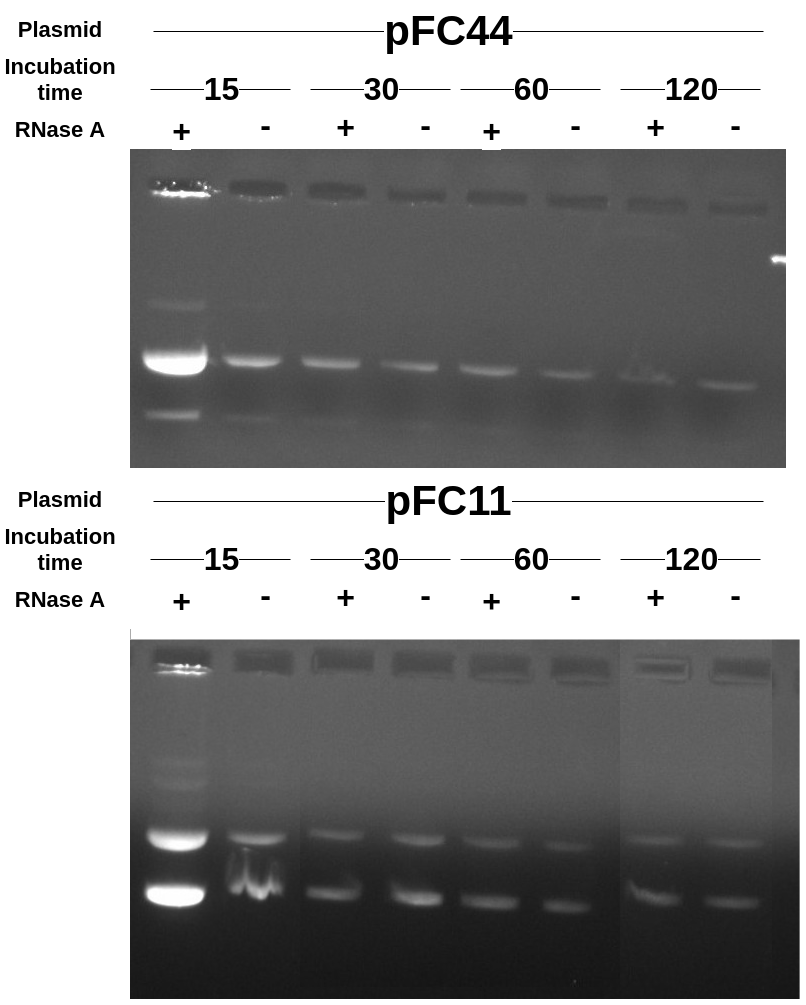
Some time dependent effect is occurring but it is observable after 15 mins and then samples look very similar. The weird part is that control samples are also being effected. This is indicating some other DNA degradation process is occurring that is independent of RNaseA activity and it is mainly observed after only 15 mins.
WTF. Fred offered DNA eating tubes as possible explanation. Everyone agreed this was probably unlikely but worth testing. Other possibilities could be effects of H20 (high acidity of npH20) so also will test effects of using DNase free H20.