Debugging IVT #
Yesterday used pFC11 and pFC8 for IVT experiments, but should have realized pFC11 is not R-loop forming. However, pFC8 did not show any transcription. I want to both retest a R-loop forming plasmid with a T7 promoter and 2 T3 R-loop formers to see if the T3 needs to be reordered.
The full FC plasmid table with respect to their promoters and R-loop forming abilities is below.
| Plasmid | RNA Pol | R-loop Forming |
|---|---|---|
| pFC1 | NA | 0 |
| pFC2 | NA | 0 |
| pFC3 | T3 | 1 |
| pFC4 | T7 | 1 |
| pFC5 | NA | 0 |
| pFC6 | T3 | 0 |
| pFC7 | T3 | 0 |
| pFC8 | T3 | 1 |
| pFC9 | T7 | 1 |
| pFC10 | T3 | 1 |
| pFC11 | T7 | 0 |
| pFC12 | NA | 0 |
| pFC13 | NA | 0 |
| pFC14 | T3 | 1 |
| pFC15 | T3 | 1 |
| pFC15 | T3 | 1 |
| pFC16 | NA | 0 |
| pFC17 | T3 | 1 |
| pFC18 | T7 | 0 |
| pFC19 | NA | 0 |
| pFC20 | T7 | 0 |
| pFC21 | T7 | 0 |
| pFC22 | T7 | 0 |
| pFC23 | T7 | 0 |
| pFC24 | T7 | 0 |
| pFC25 | T7 | 0 |
| pFC26 | T7 | 0 |
| pFC27 | T7 | 0 |
| pFC28 | T7 | 0 |
| pFC29 | T3 | 0 |
| pFC30 | NA | 0 |
| pFC31 | NA | 0 |
| pFC32 | T3 | 0 |
| pFC33 | T7 | 0 |
| pFC34 | NA | 0 |
| pFC35 | RSVLTR | 0 |
| pFC36 | RSVLTR | 0 |
| pFC37 | RSVLTR | 0 |
| pFC38 | T7 | 0 |
| pFC39 | T3 | 0 |
| pFC40 | RSV | 0 |
| pFC41 | T7 | 0 |
| pFC42 | NA | 0 |
| pFC43 | NA | 0 |
| pFC44 | T7 | 0 |
| pFC45 | T3 | 0 |
| pFC46 | T3 | 0 |
| pFC47 | RSV | 0 |
| pFC48 | RSV | 0 |
| pFC49 | RSV | 0 |
| pFC50 | T3 | 0 |
| pFC51 | T3 | 0 |
| pFC52 | T3 | 0 |
| pFC53 | T3 | 0 |
| pFC54 | CMV | 0 |
| pFC55 | CMV | 0 |
| pFC56 | RSV | 0 |
| pFC57 | RSV | 0 |
| pFC58 | NA | 0 |
| pFC59 | NA | 0 |
| pFC60 | NA | 0 |
| pFC61 | NA | 0 |
| pFC62 | NA | 0 |
| pFC63 | RSV | 0 |
| pFC64 | NA | 0 |
| pFC65 | NA | 0 |
| pFC66 | NA | 0 |
Currently have pFC8, 11, 14, 15, 17, 32 and 44 in the IVT box.
Also re-read the protocol and was using too much DTT (should be 10 mM) and way too much RNaseA. Fred says dilute 0.1mg/ml to 100x. That was probably eating up any R-loops that might have formed.
Protocol #
pFC8 DNA concentration: 240 ng / ul. pFC14 DNA concentration: 1452 ng / ul.
| Reagent | pFC8 | pFC14 |
|---|---|---|
| 5x Buffer | 4 | 4 |
| 100 mM DTT | 2 | 2 |
| 25 mM NTP | 0.4 | 0.4 |
| H20 | 11.1 | 13.187 |
| DNA | 2.5 | 0.413 |
Diluted stock RNaseA 100x and added 0.5 ul per sample. Both samples use the T3 Pol.
Run gel for 2hr at 60v, afterwards stained with EtBr for ~15 mins but added 8 ul instead of the usual 2 ul.
Gel image #
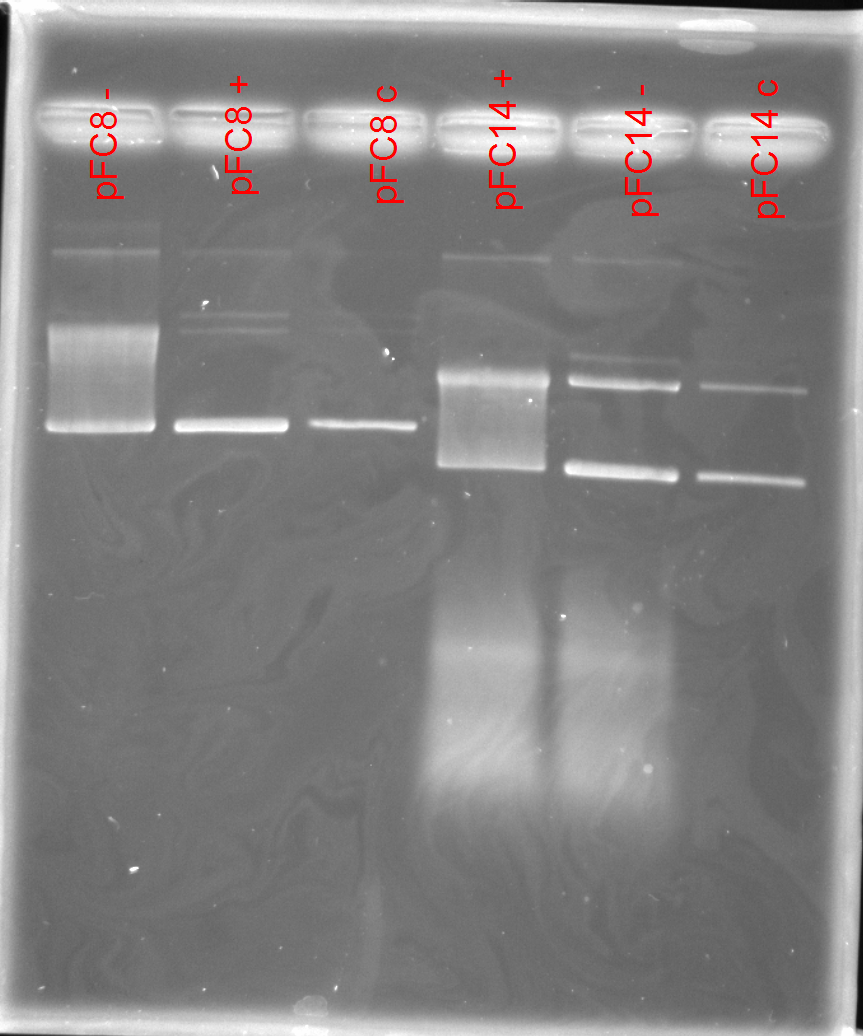
Labeling scheme #
| Symbol | Meaning |
|---|---|
| + | RNaseH containing |
| - | RNaseH control |
| c | Un-transcribed |
\d | Plasmid number |
Reagent details #
After talking with Fred agreed there was some shift but a lot of noise, the source of which could be bad reagents. Checked all reagents and multiple were expired. Also noted lot numbers, but should order all new ones tomorrow.
| Reagent | Lot Number | Expiration Date |
|---|---|---|
| RNaseH | 10016798 | 8/20 |
| DTT | 0000264878 | Not listed |
| RNA Pol T3 | 10060118 | 12/21 |
| rNTP Mix | 0291704 | 4/19 |
| 5x buffer | 004180 | 2/21 |